Search Count: 26
 |
Organism: Spiromastix sp. scsio f190
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2025-02-12 Classification: BIOSYNTHETIC PROTEIN |
 |
Structure Of Promiscuous Short Chain Dehydrogenase Spm14 In Complex With Nadph
Organism: Spiromastix sp. scsio f190
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-02-12 Classification: BIOSYNTHETIC PROTEIN Ligands: NAP, TRS |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2024-10-23 Classification: MEMBRANE PROTEIN Ligands: JAA |
 |
Cryo-Em Structure Of Jasmonic Acid Transporter Abcg16 In Occluded Conformation
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2024-10-23 Classification: MEMBRANE PROTEIN Ligands: ADP, VO4 |
 |
Cryo-Em Structure Of Jasmonic Acid Transporter Abcg16 In Outward Conformation
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2024-10-23 Classification: MEMBRANE PROTEIN Ligands: ADP, BEF, MG |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2024-10-23 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Structure Of Abscisic Acid Transporter Atabcg25 In Inward Conformation
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: MEMBRANE PROTEIN |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: MEMBRANE PROTEIN Ligands: A8S |
 |
Cryo-Em Structure Of Abscisic Acid Transporter Atabcg25 In Outward Conformation
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:3.04 Å Release Date: 2023-09-13 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: MEMBRANE PROTEIN Ligands: ATP |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: MEMBRANE PROTEIN Ligands: Y01 |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Inhibitor Gd-9
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2023-08-09 Classification: VIRAL PROTEIN Ligands: OZI, CL, BR |
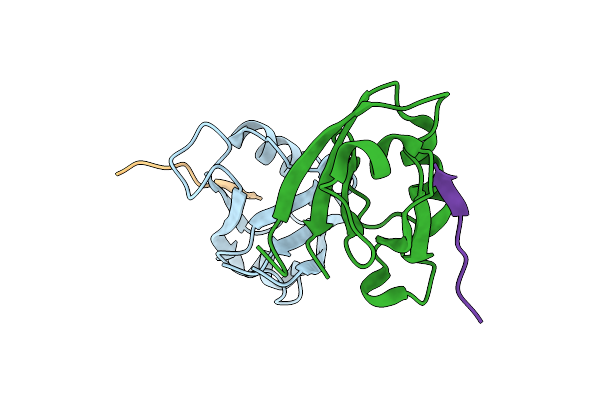 |
Crystal Structure Of The Second Pdz Domain From Human Ptpn13 In Complex With Apc Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-05-17 Classification: PEPTIDE BINDING PROTEIN |
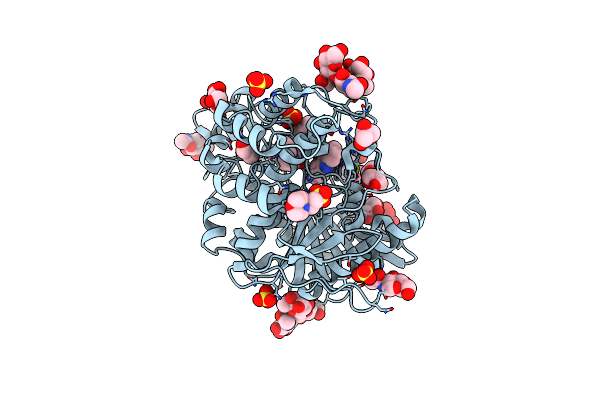 |
Human Butyrylcholinesterase In Complex With (S)-1-(4-((2-(1H-Indol-3-Yl)Ethyl)Carbamoyl)Benzyl)-N-(3-((1,2,3,4-Tetrahydroacridin-9-Yl)Amino)Propyl)Piperidine-3-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2022-12-21 Classification: HYDROLASE Ligands: NAG, MES, GOL, C0I, SIA, SO4, CL |
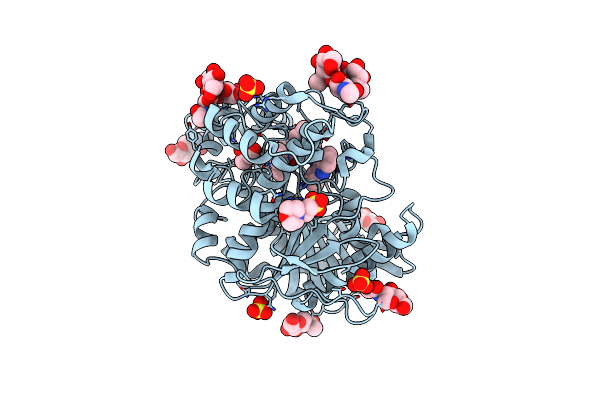 |
Human Butyrylcholinesterase In Complex With (S)-1-(4-((Naphthalen-1-Yl)Carbamoyl)Benzyl)-N-(3-((1,2,3,4-Tetrahydroacridin-9-Yl)Amino)Propyl)Piperidine-3-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2022-12-21 Classification: HYDROLASE Ligands: NAG, MES, C4I, GOL, SO4, CL |
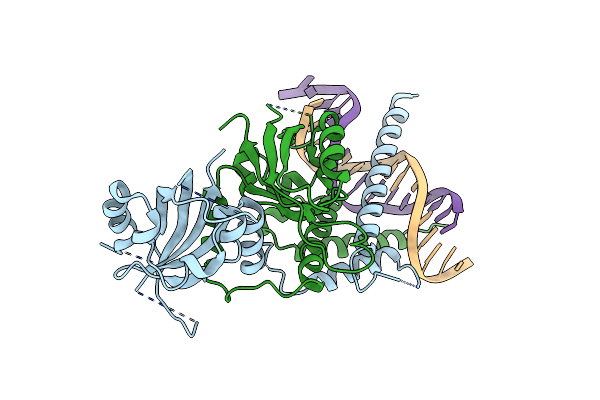 |
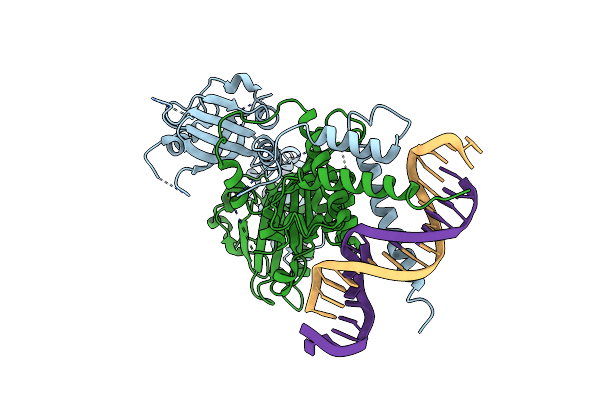
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2022-11-02 Classification: TRANSCRIPTION/DNA |
 |
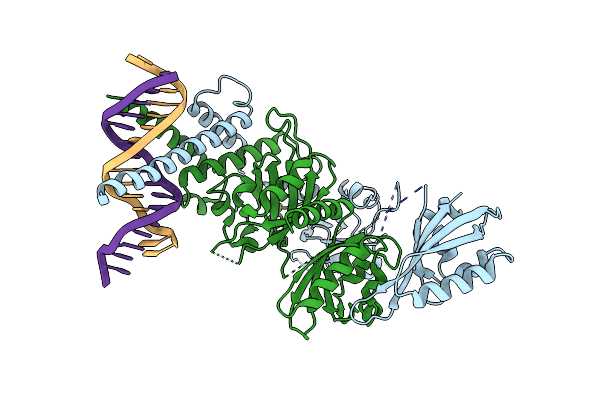
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:4.27 Å Release Date: 2022-11-02 Classification: TRANSCRIPTION/DNA |
 |
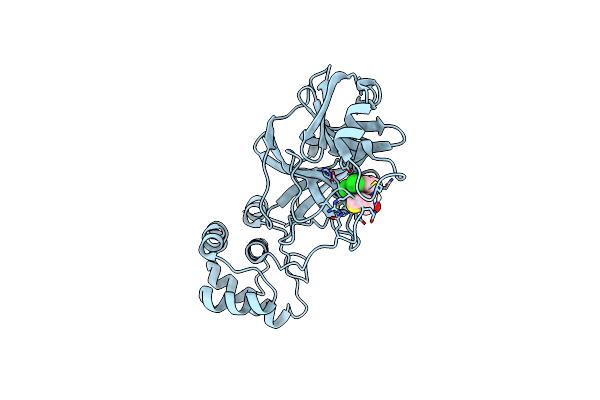
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:4.71 Å Release Date: 2022-11-02 Classification: TRANSCRIPTION |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Non-Covalent Inhibitor Ga-17S
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2022-09-28 Classification: VIRAL PROTEIN Ligands: LQ6 |

