Search Count: 43
 |
Organism: Homo sapiens, Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: GENE REGULATION Ligands: ZN |
 |
Structure Of Human Line-1 Orf2P With Endogenous Dna And Rna/Cdna Hybrid Bound To Dntp And Mn2+
Organism: Homo sapiens, Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: GENE REGULATION Ligands: DTP, ZN, MN |
 |
Structure Of Full-Length Human Line-1 Orf2P With Endogenous Dna And Rna/Cdna Hybrid
Organism: Homo sapiens, Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: GENE REGULATION Ligands: ZN |
 |
Crystal Structure Of 1-2C-T96F Tcr In Complex With Hla-A*11:01 Bound To Kras-G12V Peptide (Vvgavgvgk)
Organism: Homo sapiens, Mus musculus, Kirsten murine sarcoma virus
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: IMMUNE SYSTEM |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-01-04 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2021-12-29 Classification: CHAPERONE Ligands: GOL, CA, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-12-29 Classification: CHAPERONE Ligands: GOL, 1PE |
 |
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-08-04 Classification: HYDROLASE Ligands: ZN, FLC |
 |
Crystal Structure Of Metallo-Beta-Lactamase Imp-1 Mutant (D120E) In Complex With Citrate.
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-08-04 Classification: HYDROLASE Ligands: ZN, FLC, AE3 |
 |
Crystal Structure Of Lilrb1 D3D4 Domain In Complex With Plasmodium Rifin (Pf3D7_1373400) V2 Domain
Organism: Homo sapiens, Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2021-03-31 Classification: IMMUNE SYSTEM Ligands: NAG, SO4 |
 |
Cryoem Structure Of Lilrb1 D3D4 Domain-Inserted Antibody Mdb1 Fab In Complex With Plasmodium Rifin (Pf3D7_1373400) V2 Domain
Organism: Homo sapiens, Plasmodium falciparum (isolate 3d7)
Method: ELECTRON MICROSCOPY Release Date: 2021-03-31 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
N-Glycosylation Of Pd-1 And Glycosylation Dependent Binding Of Pd-1 Specific Monoclonal Antibody Camrelizumab
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2020-10-14 Classification: IMMUNE SYSTEM Ligands: NAG |
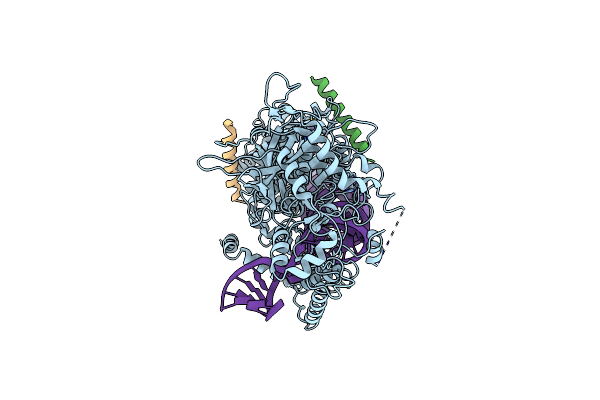 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: HYDROLASE/RNA BINDING PROTEIN/RNA Ligands: ZN |
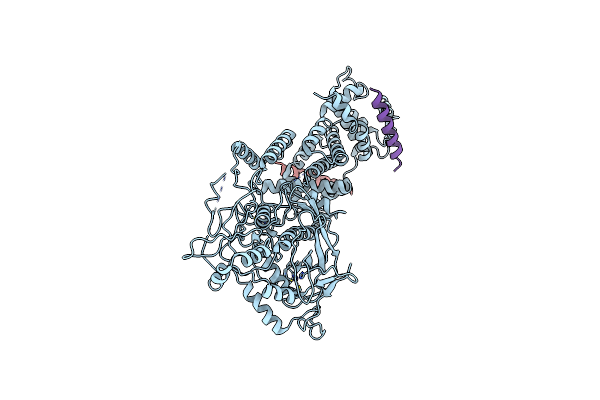 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: HYDROLASE/RNA BINDING PROTEIN Ligands: ZN |
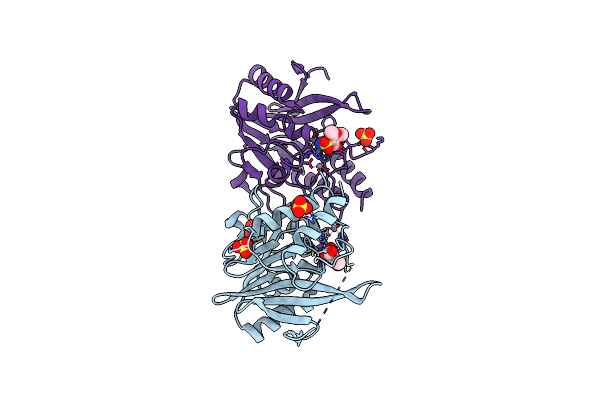 |
Crystal Structure Of Ndm-1 Metallo-Beta-Lactamase In Complex With Inhibitor No9
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2020-02-19 Classification: HYDROLASE Ligands: ZN, SO4, NO9 |
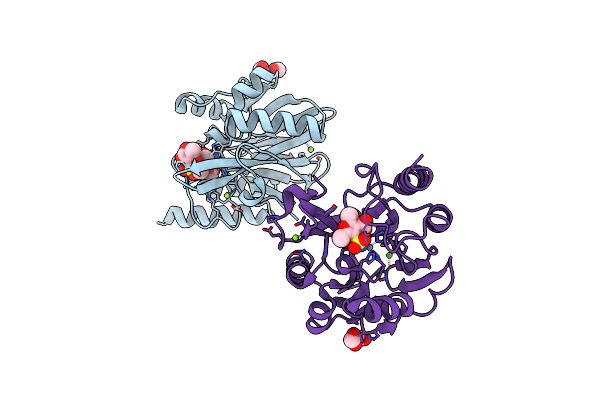 |
Crystal Structure Of Vim-2 Metallo-Beta-Lactamase In Complex With Inhibitor No9
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2020-02-19 Classification: HYDROLASE Ligands: ZN, MG, NO9, FMT |
 |
Crystal Structure Of Ndm-1 Metallo-Beta-Lactamase In Complex With Inhibitor X2
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2020-02-19 Classification: HYDROLASE Ligands: ZN, SO4, E1C |
 |
Crystal Structure Of Vim-2 Metallo-Beta-Lactamase In Complex With Inhibitor X2
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-02-19 Classification: HYDROLASE Ligands: ZN, FMT, E1C |
 |
Crystal Structure Of Imp-1 Metallo-Beta-Lactamase In Complex With No9 Inhibitor
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2020-02-19 Classification: HYDROLASE Ligands: ZN, NA, NO9 |
 |
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2019-08-07 Classification: HYDROLASE Ligands: ZN, MCR, EPE |

