Search Count: 116
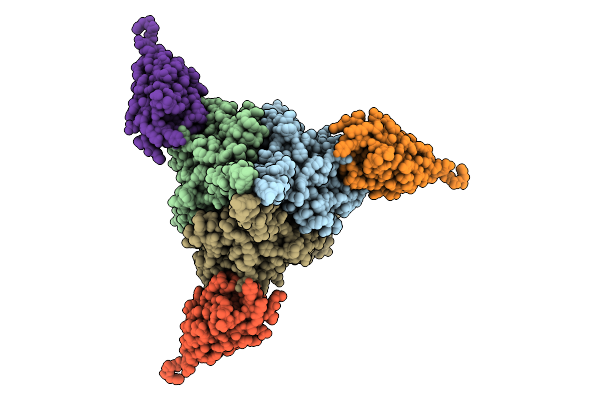 |
Cryo-Em Structure Of Prefusion-Stabilized Rsv F (Ds-Cav1 Strain: A2) In Complex With Nanobody 1G9
Organism: Respiratory syncytial virus a2, Camelus bactrianus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: ANTIVIRAL PROTEIN/IMMUNE SYSTEM |
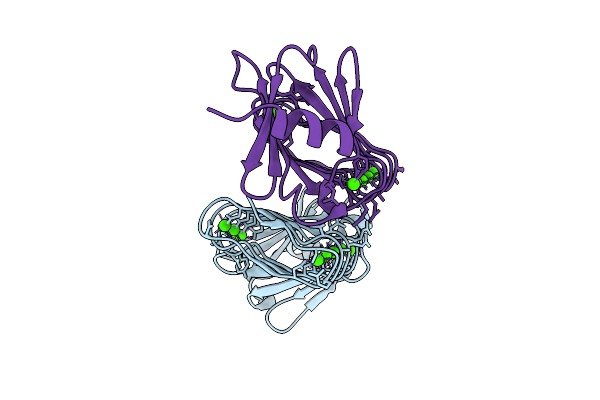 |
Rtx Domain Block V Of Adenylate Cyclase Toxin With Mutations D1533N, A1542N, D1560N, S1569N, D1587N, H1598N, H1608N
Organism: Bordetella pertussis
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2025-12-24 Classification: TOXIN Ligands: CA |
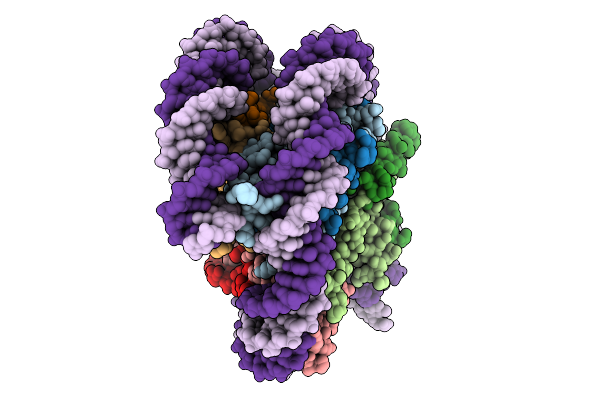 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: DNA BINDING PROTEIN/DNA |
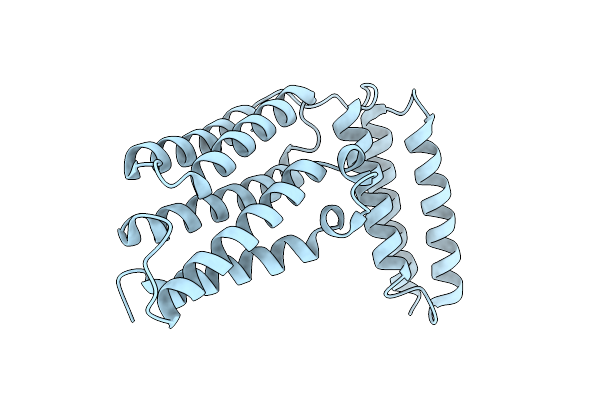 |
Organism: Cutibacterium acnes
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2025-07-23 Classification: TOXIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSFERASE/INHIBITOR Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSFERASE/INHIBITOR Ligands: AGS |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSFERASE/INHIBITOR |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSFERASE/INHIBITOR |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSFERASE/INHIBITOR Ligands: A1AEN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSFERASE/INHIBITOR Ligands: A1AEN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSFERASE/INHIBITOR |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSFERASE/INHIBITOR |
 |
Organism: Human alphaherpesvirus 2, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:3.72 Å Release Date: 2025-05-28 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Human alphaherpesvirus 2, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2025-05-28 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
Organism: Procambarus clarkii
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2025-04-09 Classification: ALLERGEN |
 |
Crystal Structure Of Sars-Cov-1 Rbd In Complex With Nanobody Asr29 And Asr347
Organism: Severe acute respiratory syndrome coronavirus, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-09-18 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
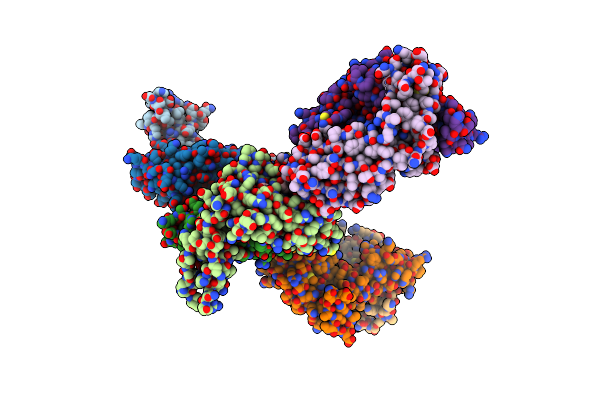 |
The Complex Of Tcr Nyn-I And Hla-A24 Bound To Sars-Cov-2 Spike448-456 Peptide Nynylyrlf
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-08-21 Classification: IMMUNE SYSTEM |
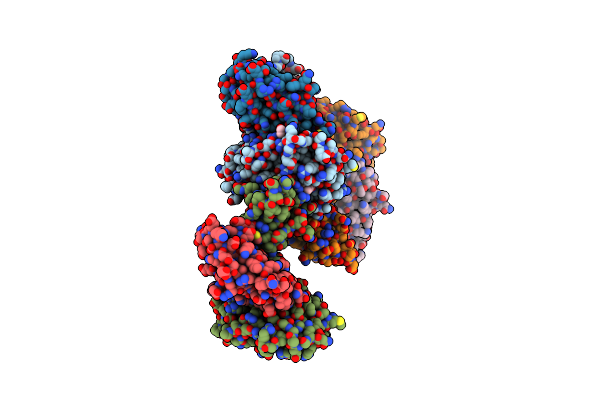 |
Complex Structure Of Hla2402 With Recognizing Sars-Cov-2 Y453F Epitope Nynylfrlf
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-08-21 Classification: IMMUNE SYSTEM Ligands: EDO |
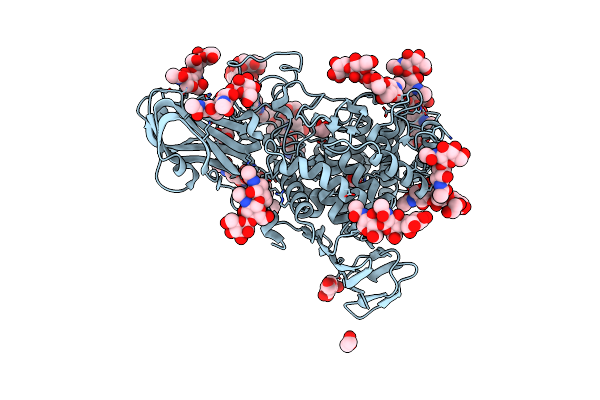 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2024-04-03 Classification: HYDROLASE Ligands: EDO, NAG |
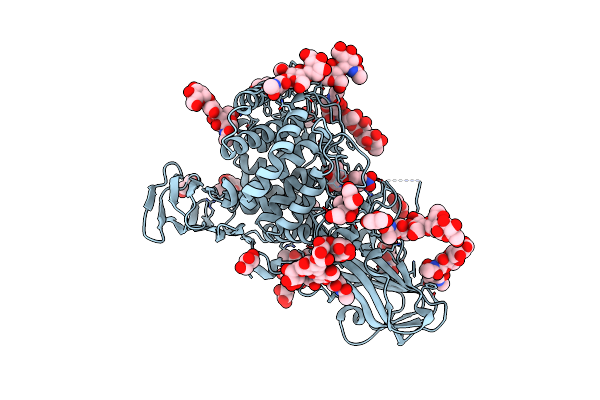 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2024-04-03 Classification: HYDROLASE Ligands: RY3, NAG, GOL |

