Search Count: 48
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: SIGNALING PROTEIN Ligands: EDO, MAN, NAG, CIT |
 |
Organism: Homo sapiens, Mus musculus, Hepatitis b virus
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Mus musculus, Hepatitis b virus
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Mus musculus, Hepatitis b virus
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
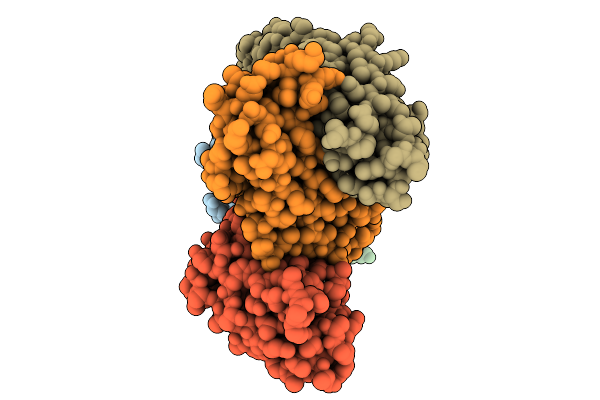 |
Organism: Homo sapiens, Lama glama, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: PEPTIDE BINDING PROTEIN/IMMUNE SYSTEM |
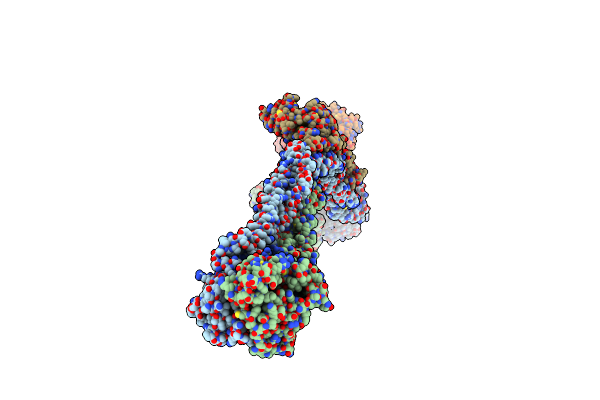 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: CELL CYCLE |
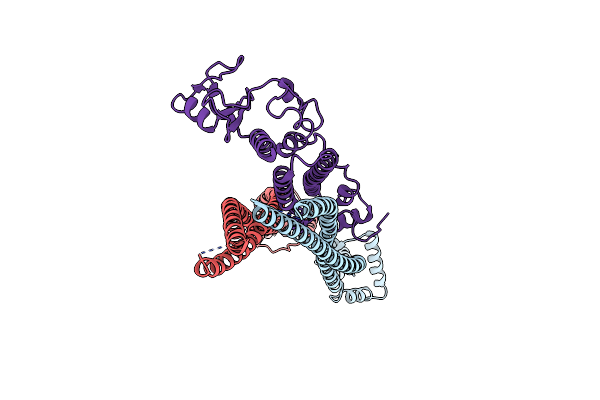 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: CELL CYCLE |
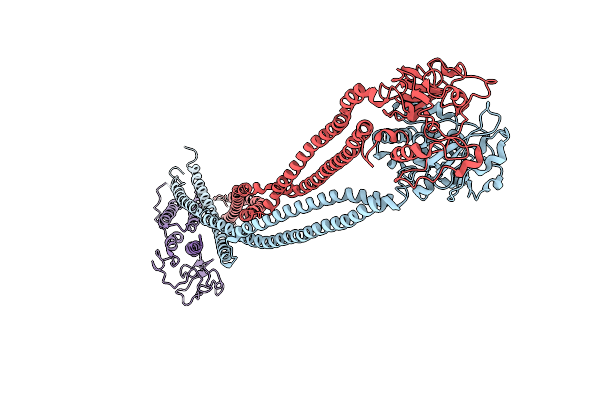 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: CELL CYCLE |
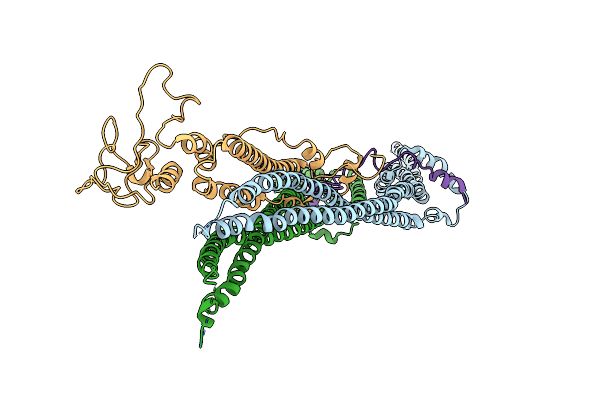 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: CELL CYCLE |
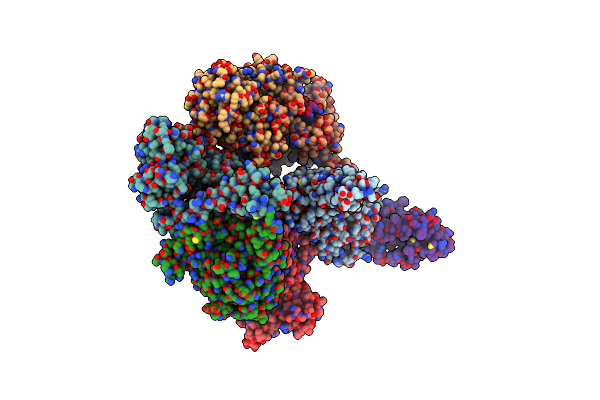 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: CELL CYCLE |
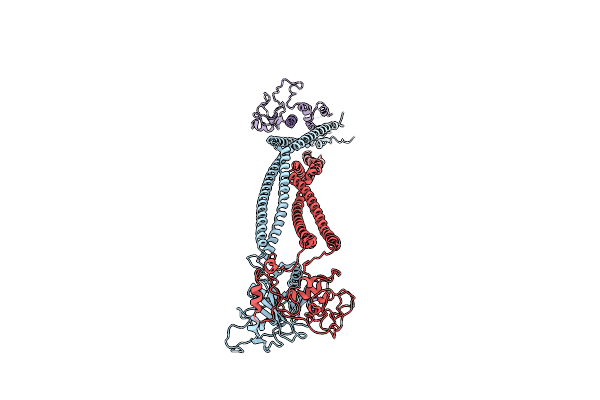 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: CELL CYCLE |
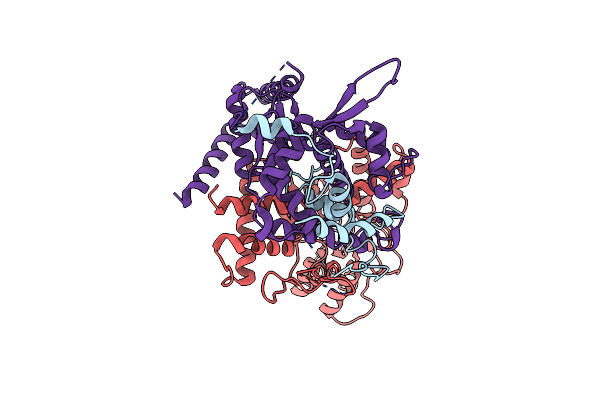 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: CELL CYCLE |
 |
Organism: Middle east respiratory syndrome-related coronavirus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-04 Classification: Ribosome/Viral Protein Ligands: MG, ZN |
 |
Crystal Structure Of Zmmcm10 In Complex With 16Nt Ssdna At 2.8. Angstrom Resolution
Organism: Zea mays, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-04-12 Classification: PLANT PROTEIN/DNA Ligands: ZN |

