Search Count: 146
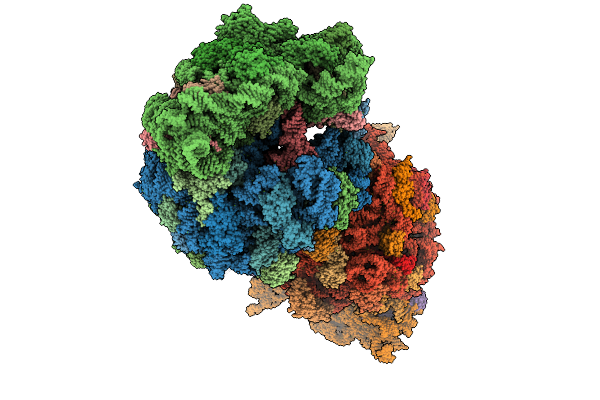 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With O-Cresomycin, Mrna, Deacylated A-Site Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.50A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: RIBOSOME Ligands: MG, K, A1A1F, ZN, SF4 |
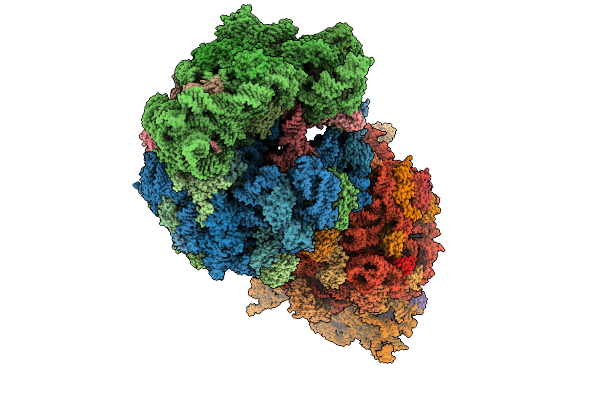 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With C-Cresomycin, Mrna, Deacylated A-Site Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.50A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: RIBOSOME Ligands: MG, K, A1A1J, ZN, SF4 |
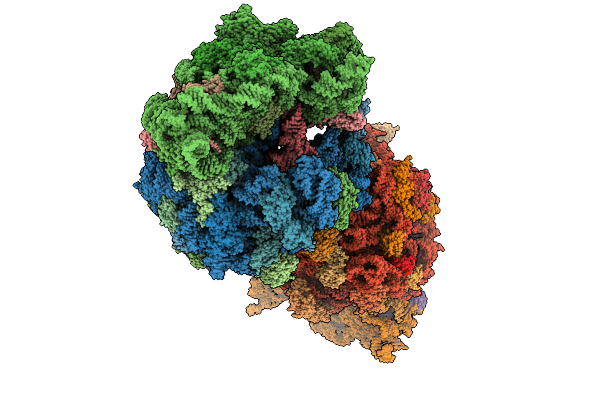 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Se-Cresomycin, Mrna, Deacylated A-Site Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.45A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: RIBOSOME Ligands: MG, K, A1A1K, ZN, SF4 |
 |
Crystal Structure Of Human Pkmyt1 Protein Kinase Domain With Naphthyridinone Inhibitor Compound 6
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: A1EJW, EDO, CL |
 |
Crystal Structure Of Human Pkmyt1 Protein Kinase Domain With Naphthyridinone Inhibitor Compound 27
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: A1EJX, SO4 |
 |
Crystal Structure Of Human Pkmyt1 Protein Kinase Domain With Naphthyridinone Inhibitor Compound 40
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2025-04-16 Classification: TRANSFERASE Ligands: A1EJU, SO4 |
 |
Crystal Structure Of Human Pkmyt1 Protein Kinase Domain With Naphthyridinone Inhibitor Compound 11
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2025-04-16 Classification: TRANSFERASE Ligands: TLA, A1EJV, EDO |
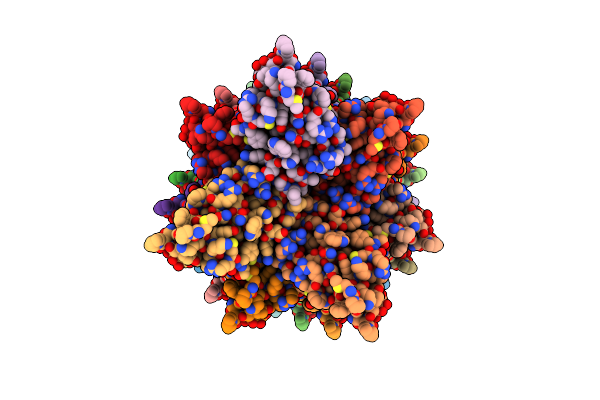 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Crispr-Csm Effector Complex From Mycobacterium Canettii
Organism: Mycobacterium canettii
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: RNA BINDING PROTEIN/RNA |
 |
Crystal Structure Of Hen Egg White Lysozyme Co-Crystallized With 10 Mm Tbxo4
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-07-17 Classification: HYDROLASE Ligands: TB, 7MT, CL, NA |
 |
Crystal Structure Of Hen Egg White Lysozyme Co-Crystallized With 10 Mm Tbxo4-Nmet2
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-07-10 Classification: HYDROLASE Ligands: TB, ZHT, CL, NA |
 |
Crystal Structure Of Hen Egg White Lysozyme Co-Crystallized With 10 Mm Tbxo4-So3
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2024-07-10 Classification: HYDROLASE Ligands: TB, ZW0, CL, NA |
 |
Crystal Structure Of Hen Egg White Lysozyme Co-Crystallized With 10 Mm Tbxo4-Oh
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-06-05 Classification: HYDROLASE Ligands: WRT, CL, NA |
 |
Organism: Mycobacterium tuberculosis, Mycobacterium canettii
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: RNA BINDING PROTEIN |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: APOPTOSIS |
 |
Organism: Schizosaccharomyces pombe 972h-
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-12-06 Classification: DNA BINDING PROTEIN Ligands: SO4 |
 |
Organism: Oryza sativa japonica group
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: ISOMERASE |
 |
Organism: Oryza sativa japonica group
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: ISOMERASE |
 |
Organism: Oryza sativa japonica group
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: ISOMERASE |
 |
Organism: Oryza sativa japonica group
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: PLANT PROTEIN Ligands: GRG |

