Search Count: 25
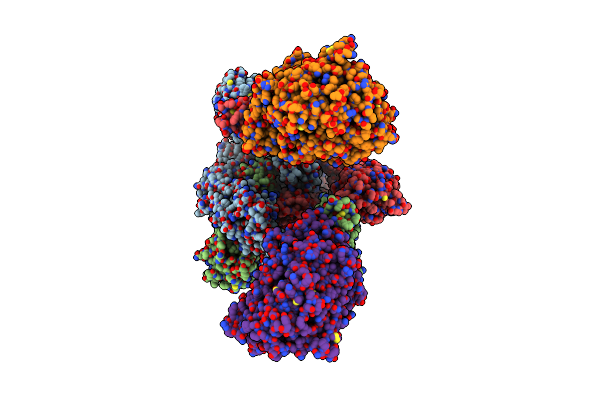 |
Cryo-Em Structure Of Mink Variant Y453F Trimeric Spike Protein Bound To Two Mink Ace2 Receptors
Organism: Severe acute respiratory syndrome coronavirus 2, Neovison vison
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: VIRAL PROTEIN |
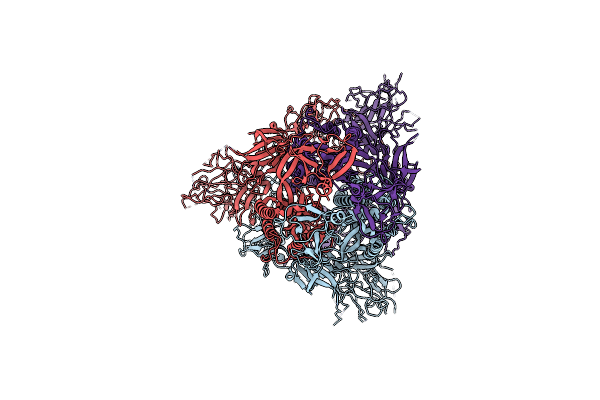 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: VIRAL PROTEIN |
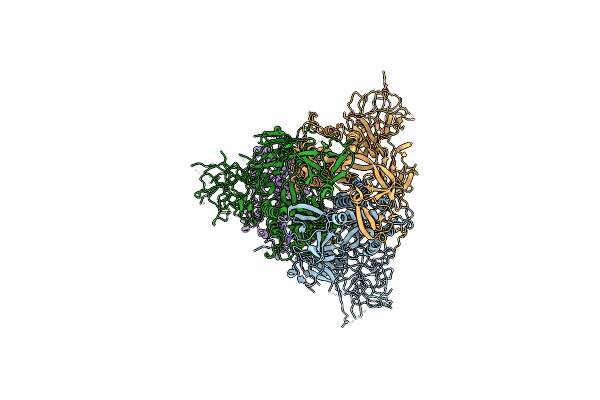 |
Cryo-Em Structure Of Mink Variant Y453F Trimeric Spike Protein Bound To One Mink Ace2 Receptors At Downrbd Conformation
Organism: Severe acute respiratory syndrome coronavirus 2, Neovison vison
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: VIRAL PROTEIN |
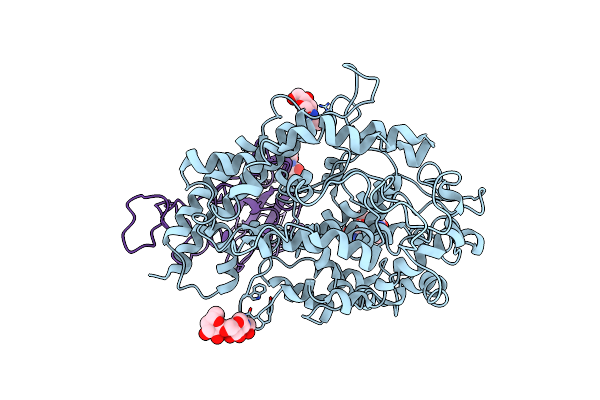 |
Cryo-Em Structure Of The Rbd-Ace2 Interface Of The Sars-Cov-2 Trimeric Spike Protein Bound To Ace2 Receptor After Local Refinement At Uprbd Conformation
Organism: Neovison vison, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of The Rbd-Ace2 Interface Of The Sars-Cov-2 Trimeric Spike Protein Bound To Ace2 Receptor After Local Refinement At Downrbd Conformation.
Organism: Neovison vison, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of Mink Variant Y453F Trimeric Spike Protein Bound To One Mink Ace2 Receptors
Organism: Severe acute respiratory syndrome coronavirus 2, Neogale vison
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: VIRAL PROTEIN |
 |
Organism: Streptomyces chartreusis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-10-26 Classification: BIOSYNTHETIC PROTEIN Ligands: FE |
 |
Organism: Streptomyces chartreusis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-05-11 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-03-03 Classification: EXOCYTOSIS Ligands: IHP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2018-01-24 Classification: METAL TRANSPORT Ligands: MLI, FE, 8R6 |
 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-07-16 Classification: Ligase/Ligase Inhibitor Ligands: 20Q, SO4 |
 |
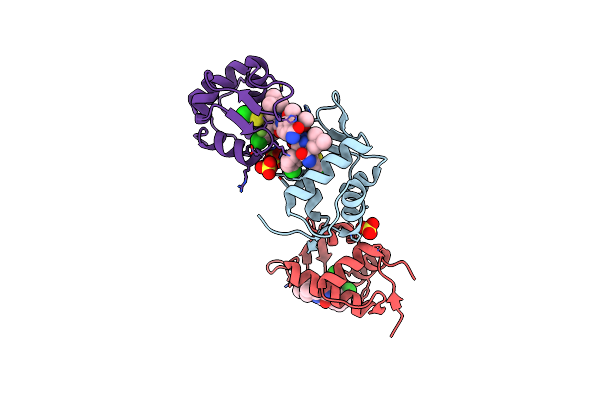
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2014-07-16 Classification: Ligase/Ligase inhibitor Ligands: 20U, SO4 |
 |
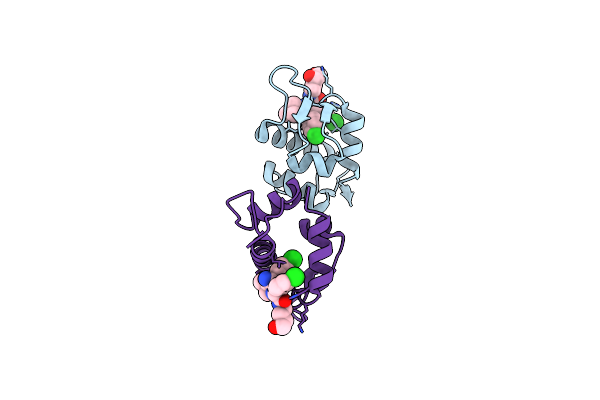
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2014-07-02 Classification: Ligase/Ligase inhibitor Ligands: 20W, SO4 |
 |
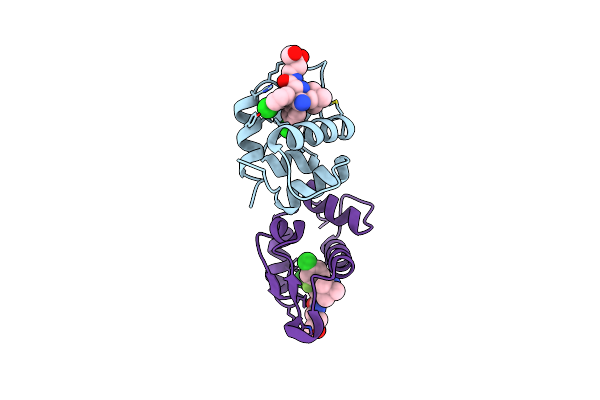
The 1.9A Crystal Structure Of Humanized Xenopus Mdm2 With Ro5313109 - A Pyrrolidine Mdm2 Inhibitor
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-07-24 Classification: LIGASE/LIGASE INHIBITOR Ligands: I09 |
 |
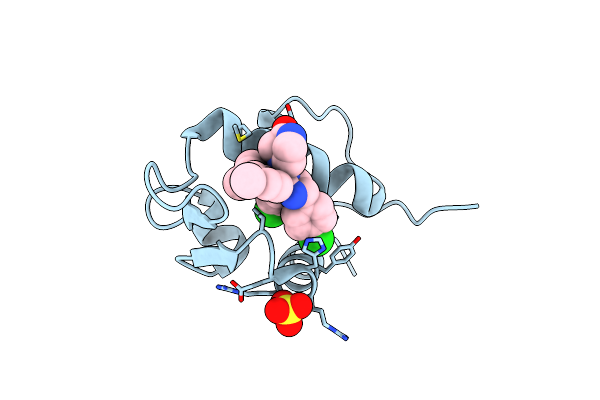
The 2.5A Crystal Structure Of Humanized Xenopus Mdm2 With Ro5316533 - A Pyrrolidine Mdm2 Inhibitor
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-07-24 Classification: LIGASE/LIGASE INHIBITOR Ligands: 1OY |
 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2013-04-24 Classification: LIGASE/ANTAGONIST Ligands: NUT, SO4 |
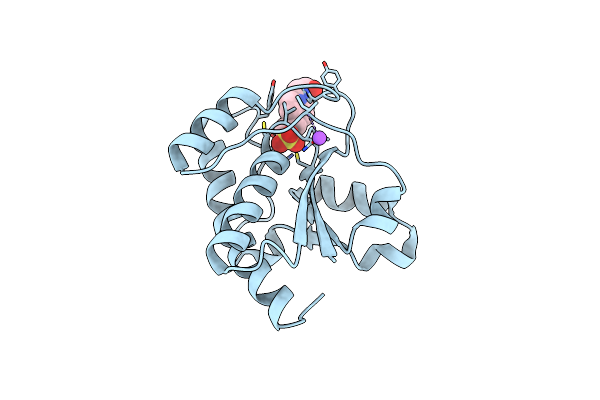 |
Crystal Structure Of Protein Tyrosine Phosphatase From Entamoeba Histolytica With Hepes In The Active Site. High Resolution, Alternative Crystal Form With 1 Molecule In Asymmetric Unit
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2009-09-22 Classification: HYDROLASE |
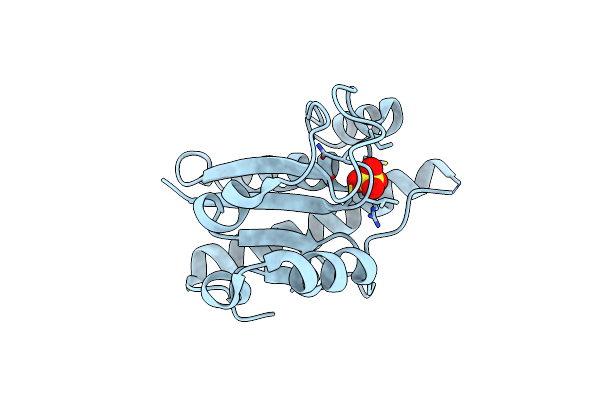 |
Product State Mimic Crystal Structure Of Protein Tyrosine Phosphatase From Entamoeba Histolytica
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-09-22 Classification: HYDROLASE Ligands: SO4 |
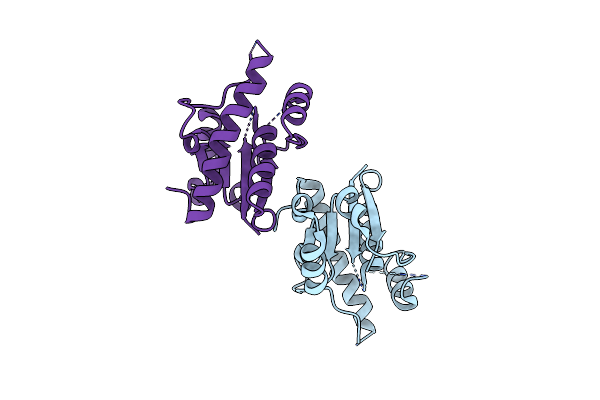 |
Apo Crystal Structure Of Protein Tyrosine Phosphatase From Entamoeba Histolytica Featuring A Disordered Active Site
Organism: Entamoeba histolytica hm-1:imss
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-08-25 Classification: HYDROLASE |
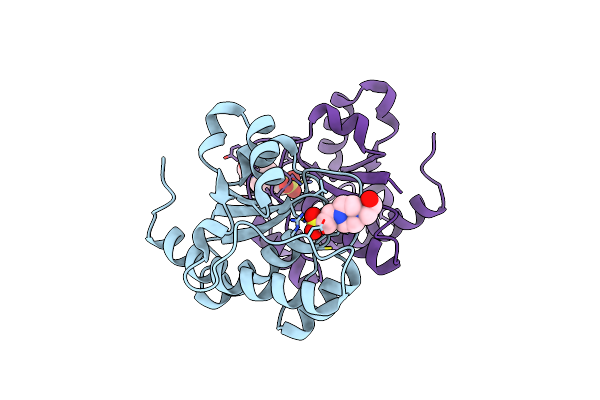 |
Crystal Structure Of Protein Tyrosine Phosphatase From Entamoeba Histolytica With A Phosphotyrosine Crude Mimic Hepes Molecule In The Active Site
Organism: Entamoeba histolytica hm-1:imss
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-07-28 Classification: HYDROLASE Ligands: EPE |

