Search Count: 177
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-10-01 Classification: METAL BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-10-01 Classification: METAL BINDING PROTEIN |
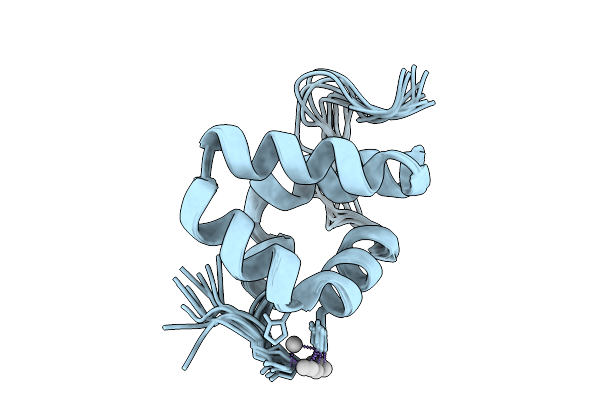 |
Solution Structure Of Silver Bound Xpc Binding Domain Of Hhr23B (Holo Form)
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-10-01 Classification: METAL BINDING PROTEIN Ligands: AG |
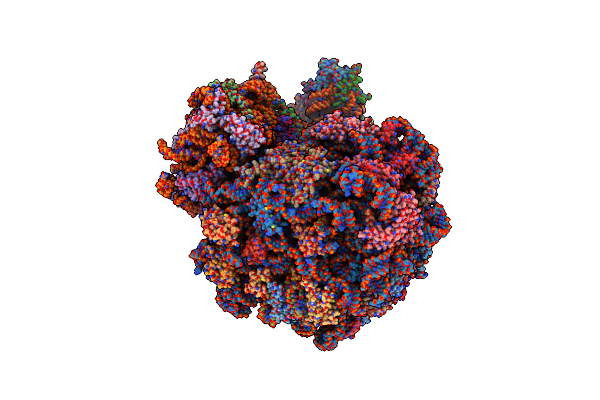 |
High-Resolution Cryo-Em Structure Of The Plasmodium Falciparum 80S Ribosome Bound To Rack1 And E-Trna
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: RIBOSOME Ligands: MG, ZN |
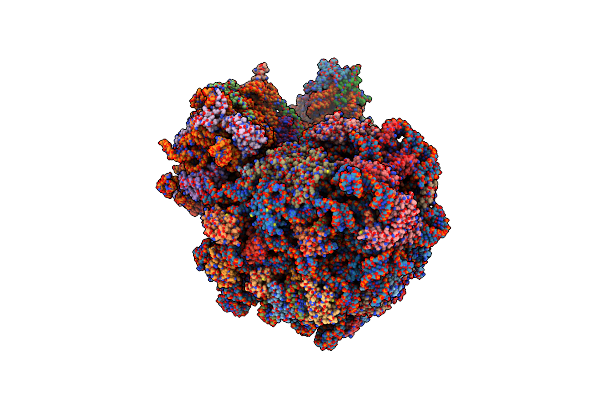 |
High-Resolution Cryo-Em Structure Of The Plasmodium Falciparum 80S Ribosome Bound To E-Trna
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: RIBOSOME Ligands: MG, ZN |
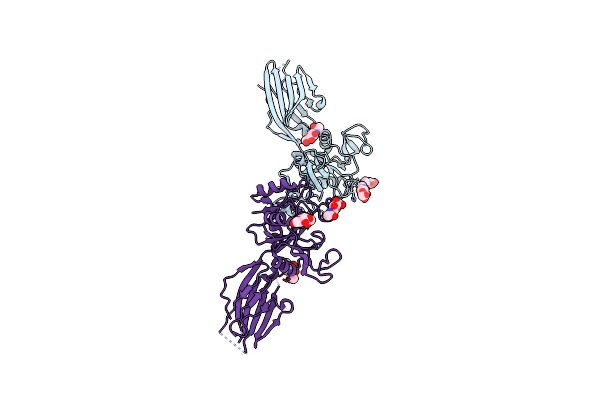 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: SUGAR BINDING PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: SUGAR BINDING PROTEIN Ligands: NAG, SO4 |
 |
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: SO4, SO3, EDO, PEG |
 |
Crystal Structure Of Nep1 In Complex With Adenosine From Pyrococcus Horikoshii Ot3
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: CL, ADN, SO4, SO3, EDO, PEG |
 |
Crystal Structure Of Nep1 In Complex With 5'-Methylthioadenosine From Pyrococcus Horikoshii Ot3
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-03-12 Classification: TRANSFERASE Ligands: CL, MTA, EDO, GOL, ACT |
 |
Crystal Structure Of Rrna (Uracil-C5)-Methyltransferase From Pyrococcus Horikoshii Ot3
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-10-16 Classification: TRANSFERASE |
 |
Cryoem Structure Of Human Full-Length Alpha1Beta3Gamma2L Gaba(A)R In Complex With Gaba And Puerarin
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: MEMBRANE PROTEIN Ligands: PGW, D10, HEX, PT5, R16, PLM, CL, ABU, NAG, A1H6W, PX2 |
 |
Crystal Structure Of Dimt1 From The Thermophilic Archaeon, Pyrococcus Horikoshii
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: ZN, SO3, GOL, PEG, EDO, ACT, ARG, EPE |
 |
Crystal Structure Of Dimt1 In Complex With 5'-Methylthioadenosine And Adenosine From Pyrococcus Horikoshii
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: MTA, ADN, ZN, CL, SO3, GOL, EDO, ACT, ARG, PEG |
 |
Crystal Structure Of Dimt1 In Complex With 5'-Methylthioadenosine From Pyrococcus Horikoshii (Formi)
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: MTA, ZN, SO3, PEG, CL, GOL, EDO |
 |
Crystal Structure Of Dimt1 In Complex With 5'-Methylthioadenosine From Pyrococcus Horikoshii (Formii)
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: MTA, ZN, SO3, GOL, EDO, PEG, PGE, ACT |
 |
Crystal Structure Of Dimt1 In Complex With Adenosylornithine (Sfg) From Pyrococcus Horikoshii
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: SFG, ZN, CL, PEG, ARG, SO3, EDO |
 |
Crystal Structure Of Dimt1 In Complex With S-Adenosyl-L-Homocysteine (Sah) From Pyrococcus Horikoshii
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: SAH, ZN, SO3, EDO, GOL, PEG, PGE, ARG |
 |
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2024-08-28 Classification: TRANSFERASE |
 |
Crystal Structure Of The D117A Mutant Of Dimt1 In Complex With 5'-Methylthioadenosine From Pyrococcus Horikoshii
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: MTA, ZN |

