Search Count: 14
 |
Organism: Rana temporaria
Method: SOLUTION NMR Release Date: 2024-06-05 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Rana temporaria
Method: SOLUTION NMR Release Date: 2024-06-05 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Rana temporaria
Method: SOLUTION NMR Release Date: 2024-06-05 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Rana temporaria
Method: SOLUTION NMR Release Date: 2023-09-06 Classification: ANTIMICROBIAL PROTEIN |
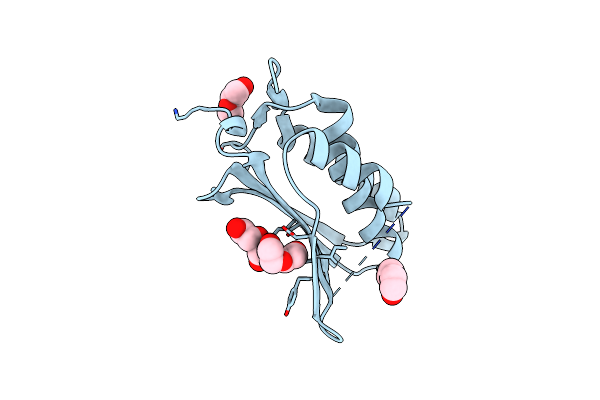 |
Crystal Structure Of The Catalytic Atp-Binding Domain Of The Phor Sensor Histidine Kinase From Vibrio Cholera
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2023-02-15 Classification: MEMBRANE PROTEIN Ligands: PEG |
 |
The 2.19-Angstrom Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Complex Minus Stalk
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: 3NI, FCO, MG, MQ9, F3S |
 |
The 1.67 Angstrom Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (Huc2S2L)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: 3NI, FCO, OH, MG, VK3, F3S |
 |
The Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Full Complex Focused Refinement Of Stalk
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: 3NI, FCO, MG, MQ9, F3S |
 |
The 1.52 Angstrom Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (Huc2S2L)
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: ELECTRON TRANSPORT Ligands: O, MG, FCO, 3NI, VK3, F3S |
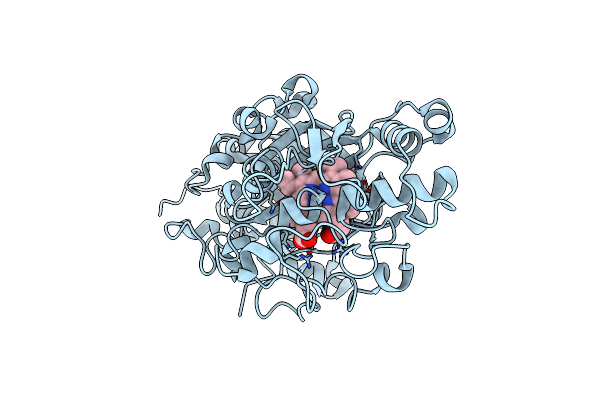 |
Organism: Irpex lacteus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-08-18 Classification: OXIDOREDUCTASE Ligands: HEM, OXY |
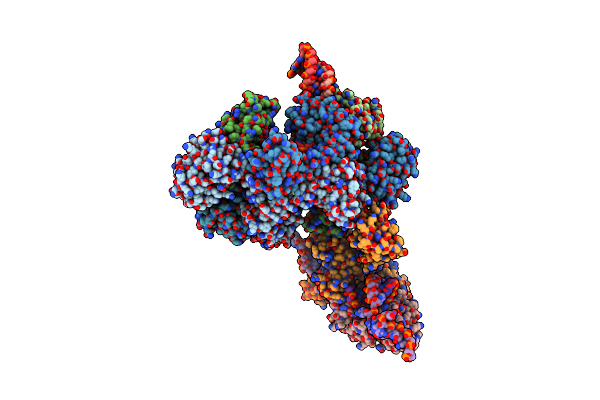 |
Crystal Structure Of A Catalytically Active Substrate-Bound Box C/D Rnp From Sulfolobus Solfataricus
Organism: Sulfolobus solfataricus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2011-01-26 Classification: TRANSFERASE/RNA Ligands: SAH |
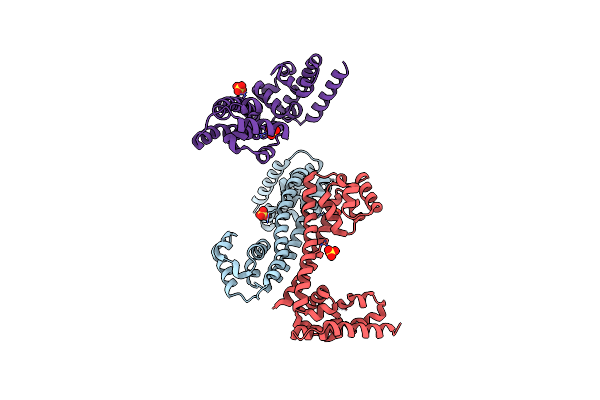 |
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2009-08-25 Classification: RNA BINDING PROTEIN Ligands: SO4 |
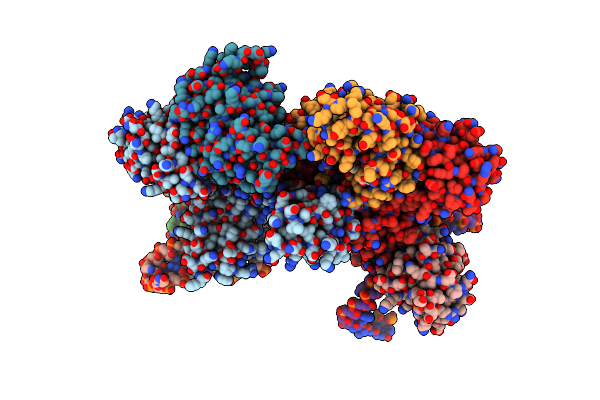 |
Crystal Structure Of Sulfolobus Solfataricus C/D Rnp Assembled With Nop5, Fibrillarin, L7Ae And A Split Half C/D Rna
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:4.01 Å Release Date: 2009-08-25 Classification: TRANSFERASE/RIBOSOMAL PROTEIN/RNA Ligands: SAM |
 |
Crystal Structure Of Sulfolobus Solfataricus Nop5 (1-262) And Fibrillarin Complex
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2009-08-25 Classification: TRANSFERASE Ligands: SAM |

