Search Count: 51
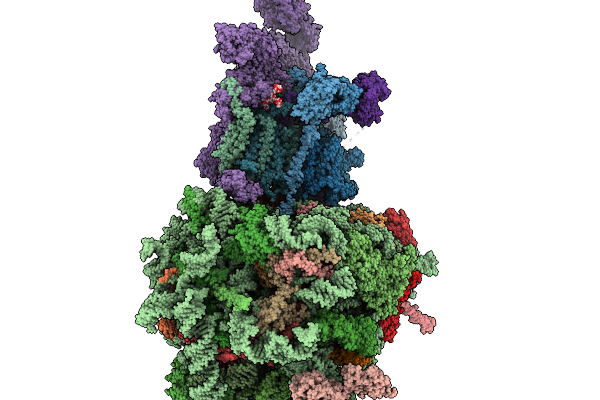 |
Structure Of A Grp94 Folding Intermediate Engaged With A Ccdc134- And Fkbp11-Bound Secretory Translocon
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: RIBOSOME Ligands: ELU, MG, ZN |
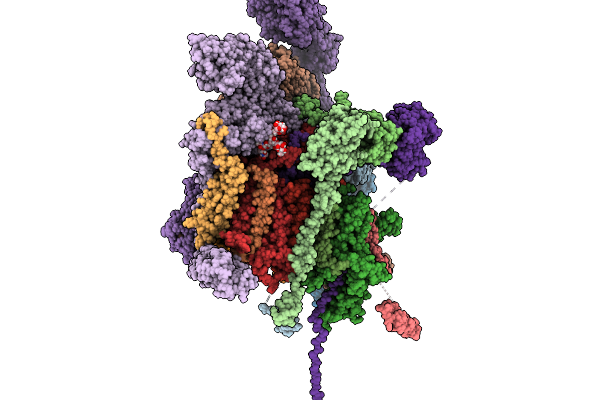 |
Structure Of A Grp94 Folding Intermediate Engaged With A Ccdc134- And Fkbp11-Bound Secretory Translocon
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: RIBOSOME Ligands: ELU |
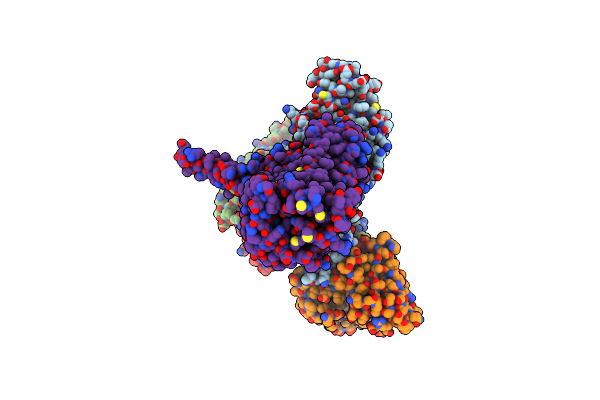 |
Organism: Homo sapiens, Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN |
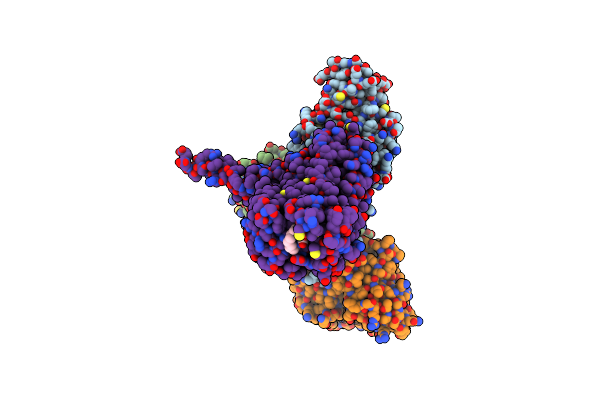 |
Organism: Homo sapiens, Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN Ligands: 140 |
 |
Organism: Homo sapiens, Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN Ligands: 9HO |
 |
Organism: Escherichia coli, Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN Ligands: NFI |
 |
Organism: Trypanosoma brucei
Method: SOLUTION NMR Release Date: 2023-03-01 Classification: RNA BINDING PROTEIN |
 |
S48A Horse Liver Alcohol Dehydrogenase In Complex With Nadh And N-Cyclohexylformamide
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-02-01 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, CXF |
 |
S48T Horse Liver Alcohol Dehydrogenase In Complex With Nadh And N-Cyclohexylformamide
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-02-01 Classification: OXIDOREDUCTASE Ligands: ZN, NAI, CXF |
 |
Cd-Substituted Horse Liver Alcohol Dehydrogenase In Complex With Nadh And N-Cyclohexylformamide
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2023-02-01 Classification: OXIDOREDUCTASE Ligands: CD, ZN, NAI, CXF |
 |
Cobalt(Ii)-Substituted Horse Liver Alcohol Dehydrogenase In Complex With Nadh And N-Cyclohexylformamide
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-02-01 Classification: OXIDOREDUCTASE Ligands: CO, ZN, NAI, CXF |
 |
Cobalt(Ii)-Substituted S48A Horse Liver Alcohol Dehydrogenase In Complex With Nadh And N-Cyclohexylformamide
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2023-02-01 Classification: OXIDOREDUCTASE Ligands: CO, ZN, NAI, CXF |
 |
Cobalt(Ii)-Substituted S48T Horse Liver Alcohol Dehydrogenase In Complex With Nadh And N-Cyclohexylformamide
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2023-02-01 Classification: OXIDOREDUCTASE Ligands: CO, ZN, NAI, CXF |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2022-09-07 Classification: HYDROLASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2022-09-07 Classification: HYDROLASE/INHIBITOR Ligands: NXL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2022-09-07 Classification: HYDROLASE/INHIBITOR Ligands: NXL |
 |
Tem-1 Beta-Lactamase A237Y Mutant Covalently Bound To Avibactam, A Room Temperature Structure
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2022-09-07 Classification: HYDROLASE/INHIBITOR Ligands: NXL |
 |
Horse Liver Alcohol Dehydrogenase In Complex With Nadh And N-Cylcohexyl Formamide
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2022-04-27 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: ZN, NAI, CXF |
 |
Structure And Mutation Analysis Of The Hexameric P4 From Pseudomonas Aeruginosa Phage Phiyy
Organism: Pseudomonas phage phiyy
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2022-03-23 Classification: HYDROLASE |

