Search Count: 156
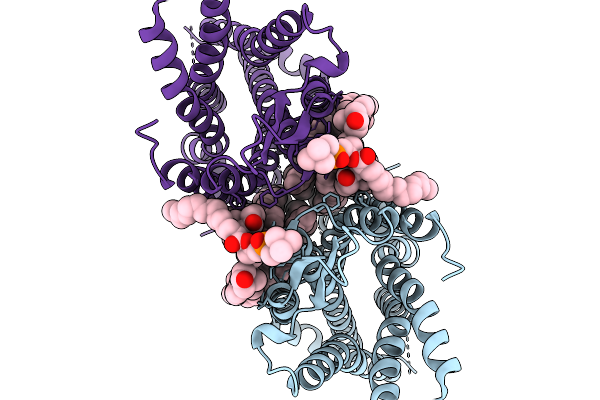 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: STRUCTURAL PROTEIN Ligands: PCF, CLR |
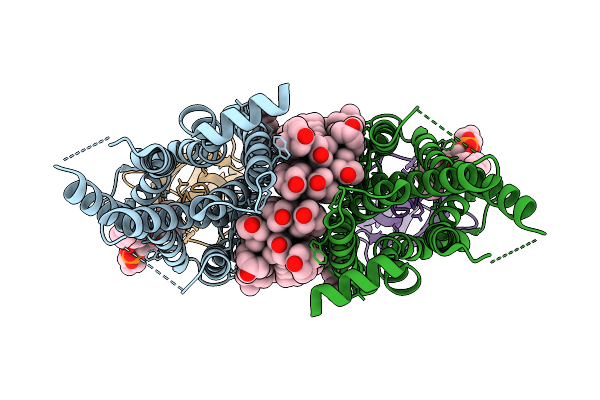 |
Organism: Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: STRUCTURAL PROTEIN Ligands: CLR, PCF |
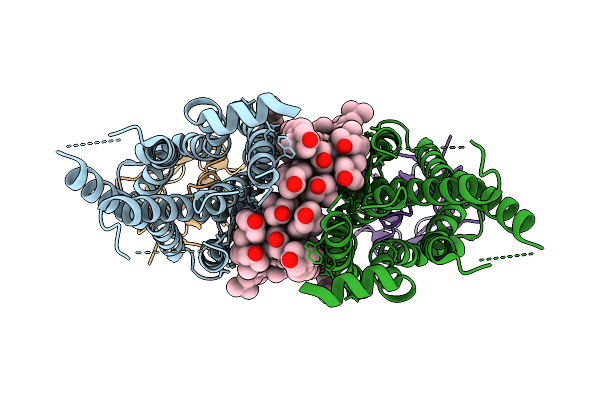 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.70 Å Release Date: 2026-02-04 Classification: STRUCTURAL PROTEIN Ligands: PCF, CLR |
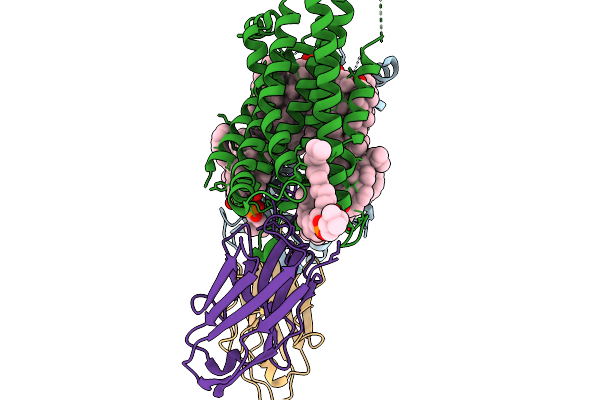 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.70 Å Release Date: 2026-02-04 Classification: STRUCTURAL PROTEIN Ligands: CLR, PCF |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: STRUCTURAL PROTEIN Ligands: PCF, A1EK3, CLR |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.60 Å Release Date: 2026-02-04 Classification: STRUCTURAL PROTEIN Ligands: A1EK3, PCF, CLR |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.50 Å Release Date: 2026-02-04 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2026-02-04 Classification: STRUCTURAL PROTEIN Ligands: PCF, CLR |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2026-02-04 Classification: STRUCTURAL PROTEIN Ligands: A1EK3 |
 |
Organism: Bacteroidales bacterium
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: IMMUNE SYSTEM/RNA Ligands: MG |
 |
Organism: Bacteroidales, Bacteroidales bacterium
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: IMMUNE SYSTEM/RNA Ligands: MG, ZN |
 |
Crystal Structure Of C36S Mutant Glutathione Peroxidase Of Staphylococcus Aureus.
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-12-17 Classification: OXIDOREDUCTASE |
 |
Organism: Homo sapiens, Oplophorus gracilirostris, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: HOH |
 |
Organism: Mus musculus, Homo sapiens, Bos taurus, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
 |
Organism: Mus musculus, Homo sapiens, Bos taurus, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
 |
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2025-11-19 Classification: FLAVOPROTEIN Ligands: FMN |
 |
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2025-11-19 Classification: FLAVOPROTEIN Ligands: FAD |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2025-09-24 Classification: ELECTRON TRANSPORT |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-09-24 Classification: ELECTRON TRANSPORT |

