Search Count: 193
 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-02-19 Classification: BIOSYNTHETIC PROTEIN Ligands: A1D52, PEG |
 |
Sfx Structure Of An Mn-Carbonyl Complex Immobilized In Hen Egg White Lysozyme Microcrystals, 100 Ns After Photoexcitation At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-09-04 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
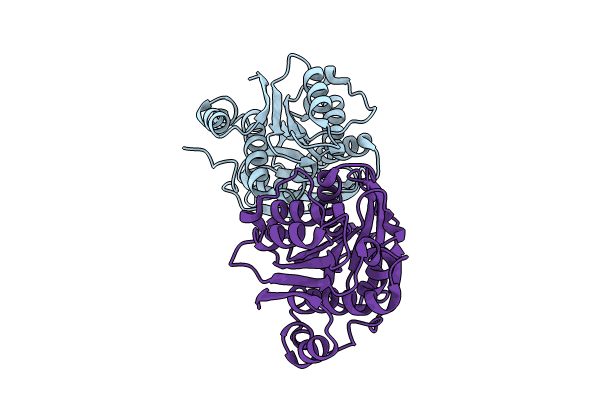
Crystal Structure Of Saccharomyces Cerevisiae Nmd4 Protein Bound To Upf1 Helicase Domain
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-08-14 Classification: RNA BINDING PROTEIN Ligands: EDO, SIN, FMT, MLA |
 |
Crystal Structure Of Saccharomyces Cerevisiae Nmd4 Protein Involved In Nonsense Mediated Mrna Decay
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-08-14 Classification: RNA BINDING PROTEIN Ligands: GOL, ACY |
 |
Dark State Sfx Structure Of Hen Egg White Lysozyme Microcrystals Immobilized With A Mn Carbonyl Complex At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
Sfx Structure Of An Mn-Carbonyl Complex Immobilized In Hen Egg White Lysozyme Microcrystals, 10 Ns After Photoexcitation At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
Sfx Structure Of An Mn-Carbonyl Complex Immobilized In Hen Egg White Lysozyme Microcrystals, 1 Microsecond After Photoexcitation At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
Sfx Structure Of An Mn-Carbonyl Complex Immobilized In Hen Egg White Lysozyme Microcrystals, 1 Microsecond After Photoexcitation With 40 Microjoules Laser Intensity At Rt.
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: CL, NA, XTX |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: DNA BINDING PROTEIN |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: DNA BINDING PROTEIN |
 |
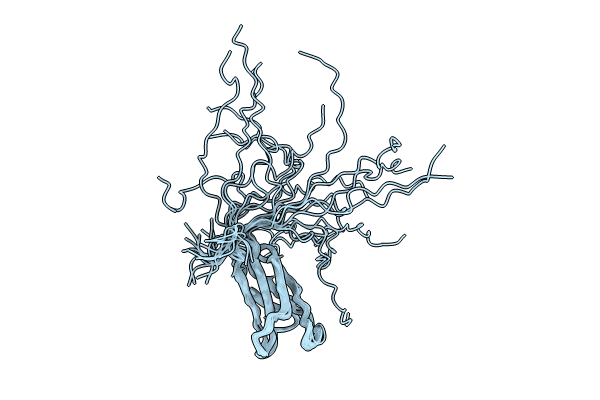
Focused Cryo-Em Map Of Msdps2 From Msdps2-Dna Complex Of Mycobacterium Smegmatis
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: DNA BINDING PROTEIN |
 |
Organism: Pyricularia oryzae 70-15
Method: SOLUTION NMR Release Date: 2024-02-14 Classification: UNKNOWN FUNCTION |
 |
Organism: Pseudomonas
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-02-14 Classification: HYDROLASE |
 |
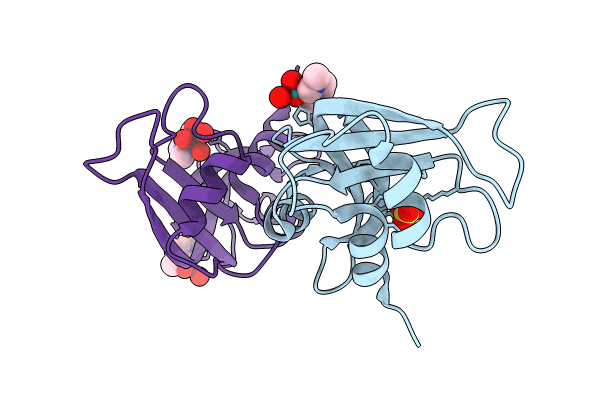
X-Ray Structure Of Hewl Upon Reaction With A Ruthenium(Ii)-Arene Complexed With Glycosylated Carbene Ligands (5)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2024-01-10 Classification: HYDROLASE |
 |
X-Ray Structure Of Rnase A Upon Reaction With A Ruthenium(Ii)-Arene Complexed With Glycosylated Carbene Ligands (5)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2024-01-10 Classification: RNA BINDING PROTEIN Ligands: SO4, R6U, T9L, T9U |
 |
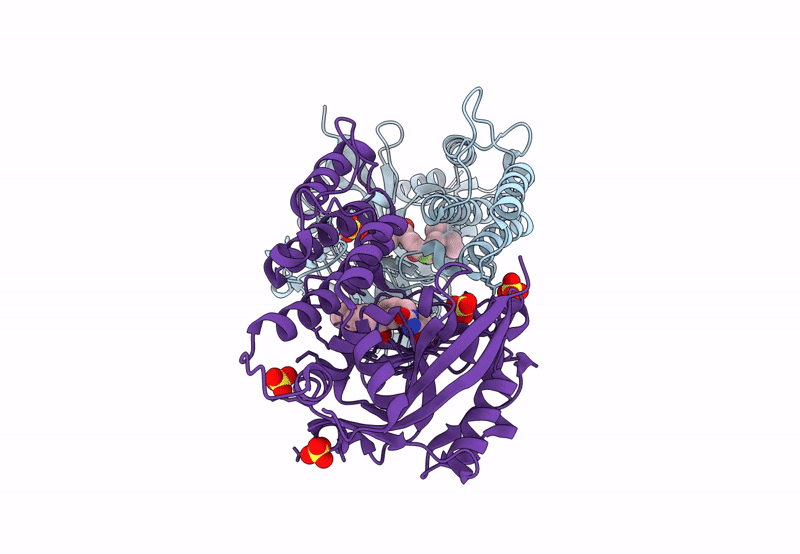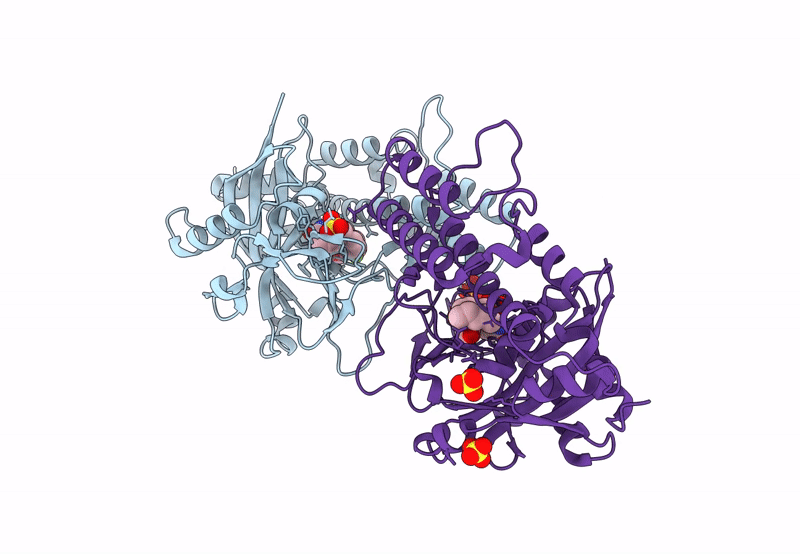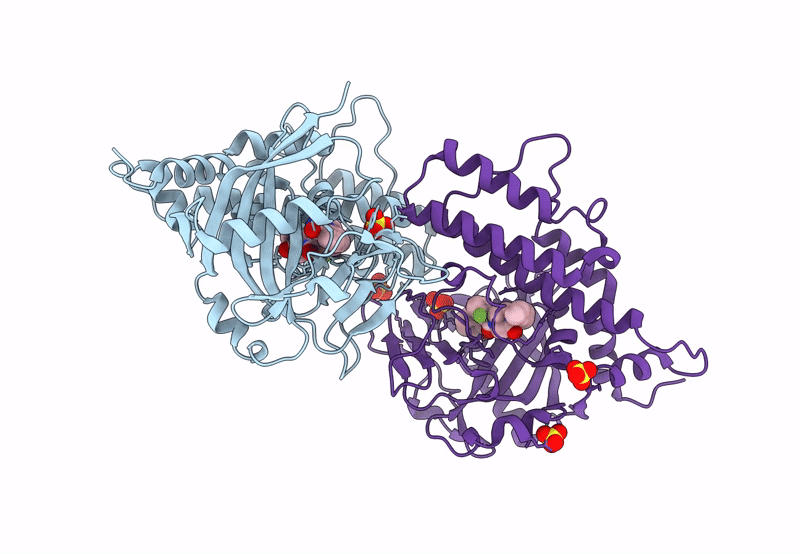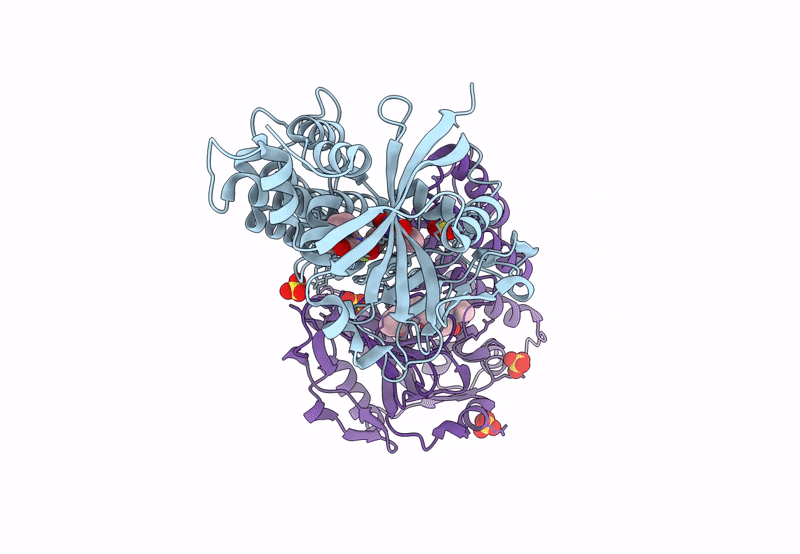
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-12-13 Classification: CELL CYCLE Ligands: XSE |
 |
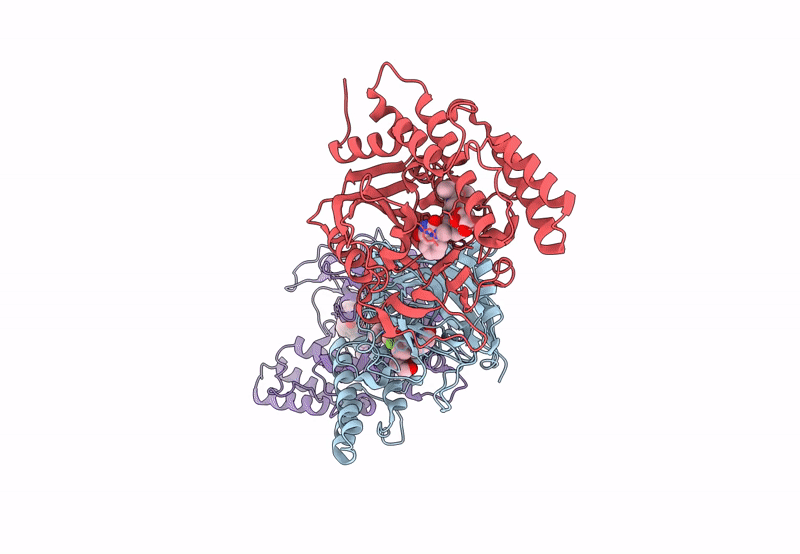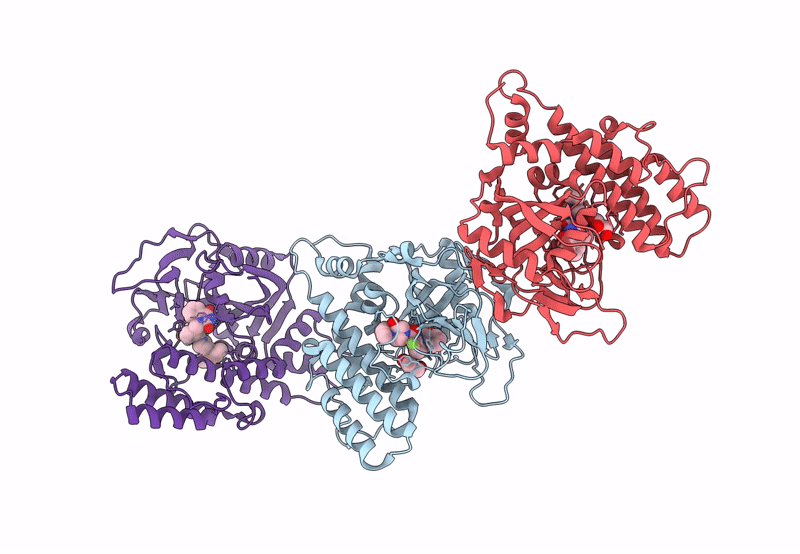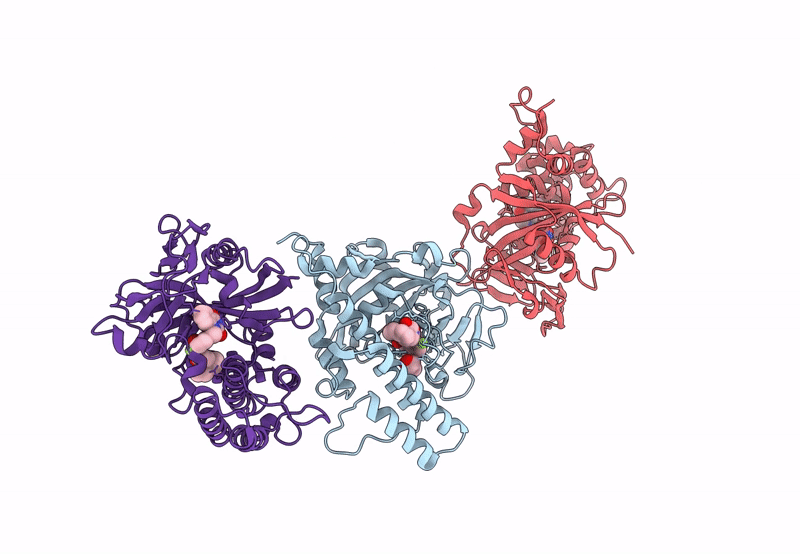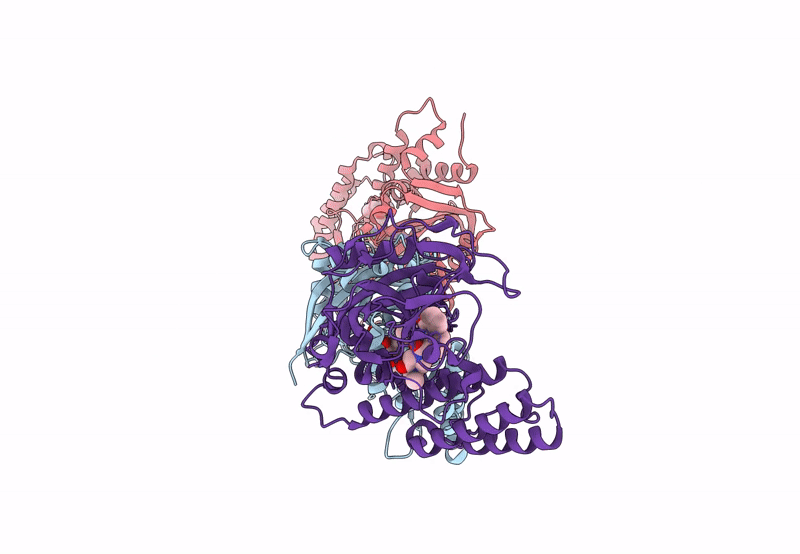
Adp-Ribosyltransferase 1 (Parp1) Catalytic Domain Bound To A Quinazoline-2,4(1H,3H)-Dione Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-11-08 Classification: TRANSFERASE Ligands: 1WI, SO4 |
 |
Human Adp-Ribosyltransferase 2 (Parp2) Catalytic Domain Bound To A Quinazoline-2,4(1H,3H)-Dione Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2023-11-08 Classification: TRANSFERASE Ligands: GOL, 1WI |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-10-04 Classification: TRANSFERASE Ligands: POV, AV0, ERG |

