Search Count: 128
 |
Structure Of E. Coli Dihydrofolate Reductase (Dhfr) In An Occluded Conformation And In Complex With A Cycloguanil Derivative
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: A1A3E |
 |
Structure Of E. Coli Dihydrofolate Reductase (Dhfr) In An Occluded Conformation And In Complex With Cycloguanil
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: 1CY, FLC |
 |
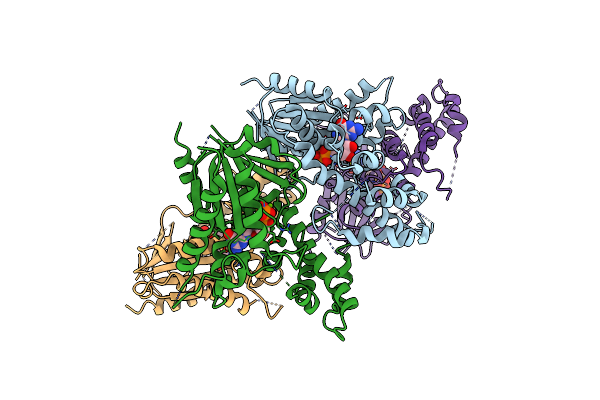
Crystal Structure Of Guanine Nucleotide-Binding Protein (G Protein) Alpha-1 Subunit From Selaginella Moellendorffii In Complex With Gtp Gamma S
Organism: Selaginella moellendorffii
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2024-11-06 Classification: SIGNALING PROTEIN Ligands: MG, GSP |
 |
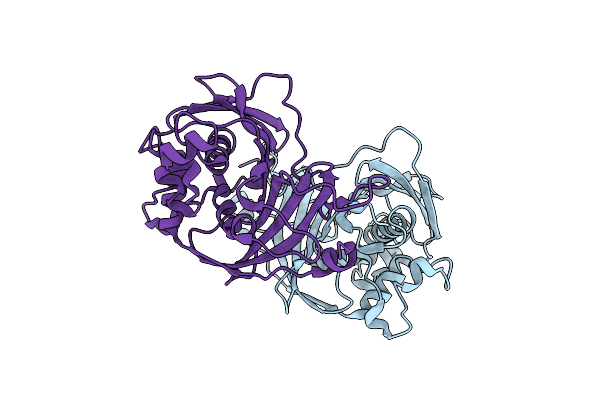
Crystal Structure Of Guanine Nucleotide-Binding Protein Alpha Subunit (G Protein) From Oryza Sativa In Complex With Gdp
Organism: Oryza sativa indica group
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2024-11-06 Classification: SIGNALING PROTEIN Ligands: GDP, MG |
 |
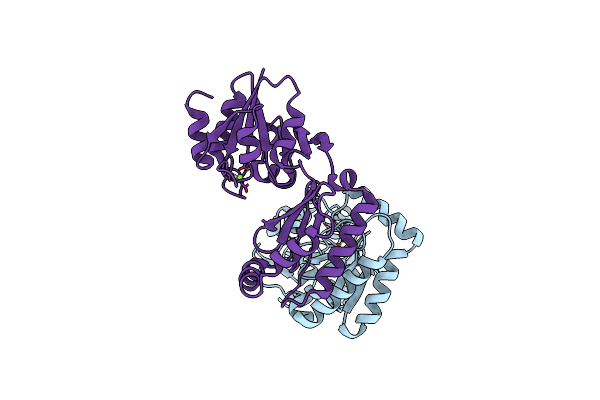
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2022-05-11 Classification: LYASE |
 |
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:3.51 Å Release Date: 2022-03-30 Classification: HYDROLASE Ligands: MG |
 |
Crystal Structure Of Mannitol Dehydrogenase (Chmdh) From Cladosporium Herbarum In Complex With Nadp+ And Na
Organism: Cladosporium herbarum
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2022-02-23 Classification: OXIDOREDUCTASE |
 |
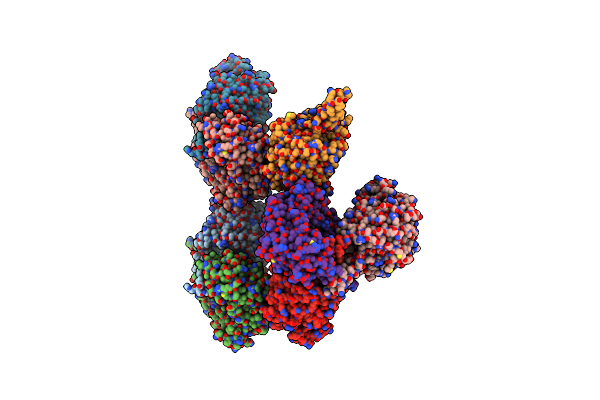
Crystal Structure Of Aldehyde Dehydrogenase (Chaldh) From Cladosporium Herbarum
Organism: Cladosporium herbarum
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2021-11-24 Classification: OXIDOREDUCTASE |
 |
P. Syringae Alda Indole-3-Acetaldehyde Dehydrogenase C302A Mutant In Complex With Nad+ And Iaa
Organism: Pseudomonas syringae pv. tomato (strain atcc baa-871 / dc3000)
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2021-06-23 Classification: OXIDOREDUCTASE Ligands: NAI, IAC |
 |
Crystal Structure Of Hydroxyacyl Glutathione Hydrolase (Glob) From Staphylococcus Aureus, Apoenzyme
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-12-30 Classification: HYDROLASE Ligands: ZN, SO4 |
 |
Crystal Structure Of S-Formylglutathione Hydrolase (Frmb) From Staphylococcus Aureus, Apoenzyme
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-12-23 Classification: HYDROLASE Ligands: MG |
 |
Crystal Structure Of Aldehyde Dehydrogenase C (Aldc) Mutant (C291A) From Pseudomonas Syringae In Complexed With Nad+ And Octanal
Organism: Pseudomonas syringae pv. tomato (strain atcc baa-871 / dc3000)
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2020-09-09 Classification: OXIDOREDUCTASE Ligands: OYA, NAD |
 |
Phosphoethanolamine Methyltransferase From The Pine Wilt Nematode Bursaphelenchus Xylophilus
Organism: Bursaphelenchus xylophilus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-06-17 Classification: TRANSFERASE Ligands: OPE, SAH |
 |
Crystal Structure Of 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase From Staphylococcus Schleiferi, Apoenzyme
Organism: Staphylococcus schleiferi
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Crystal Structure Of 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase From Staphylococcus Schleiferi In Complex With Fosmidomycin (Fom)
Organism: Staphylococcus schleiferi
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: FOM |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2019-10-09 Classification: LIGASE Ligands: AMP, ISJ |
 |
Crystal Structure Of Chalcone Isomerase From Medicago Truncatula Complexed With (2S) Naringenin
Organism: Medicago truncatula
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-08-21 Classification: plant protein, isomerase Ligands: NAR |
 |
Crystal Structure Of Semet Udp-Dependent Glucosyltransferases (Ugt) From Stevia Rebaudiana In Complex With Udp
Organism: Stevia rebaudiana
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-06-12 Classification: TRANSFERASE Ligands: UDP |
 |
Crystal Structure Of Udp-Dependent Glucosyltransferases (Ugt) From Stevia Rebaudiana In Complex With Udp
Organism: Stevia rebaudiana
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-06-12 Classification: TRANSFERASE Ligands: UDP |
 |
Crystal Structure Of Udp-Dependent Glucosyltransferases (Ugt) From Stevia Rebaudiana In Complex With Udp And Rebaudioside A
Organism: Stevia rebaudiana
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2019-06-12 Classification: TRANSFERASE Ligands: UDP, LRV |

