Search Count: 39
 |
Organism: Homo sapiens, Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: CELL CYCLE |
 |
Organism: Synthetic construct, Xenopus laevis, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.86 Å Release Date: 2025-10-01 Classification: CELL CYCLE |
 |
Organism: Xenopus laevis, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.85 Å Release Date: 2025-10-01 Classification: CELL CYCLE |
 |
Chromosomal Passenger Complex In Complex With H3T3Ph Nucleosome (Double Occupancy)
Organism: Homo sapiens, Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.80 Å Release Date: 2025-10-01 Classification: CELL CYCLE |
 |
Organism: Oryza, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: DNA BINDING PROTEIN |
 |
Organism: Oryza, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2024-10-23 Classification: CELL CYCLE Ligands: SAH |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2024-10-23 Classification: CELL CYCLE Ligands: SAM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-10-23 Classification: CELL CYCLE Ligands: HOH |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-08-21 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2024-08-21 Classification: CELL CYCLE |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:4.36 Å Release Date: 2020-04-15 Classification: CELL CYCLE |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:3.47 Å Release Date: 2020-04-01 Classification: CELL CYCLE |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2020-04-01 Classification: CELL CYCLE |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2020-04-01 Classification: CELL CYCLE |
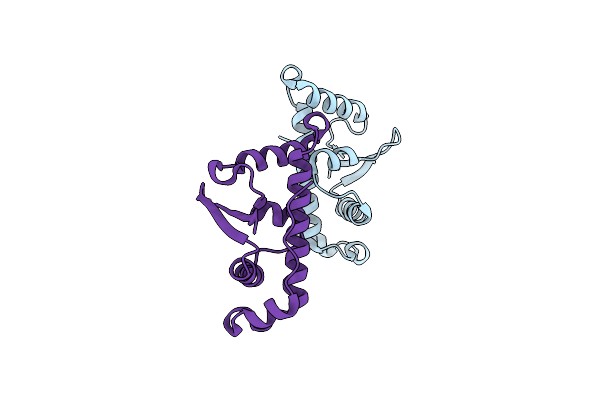 |
Structural Basis For The Microtubule Binding Of The Human Kinetochore Ska Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2014-01-22 Classification: CELL CYCLE |
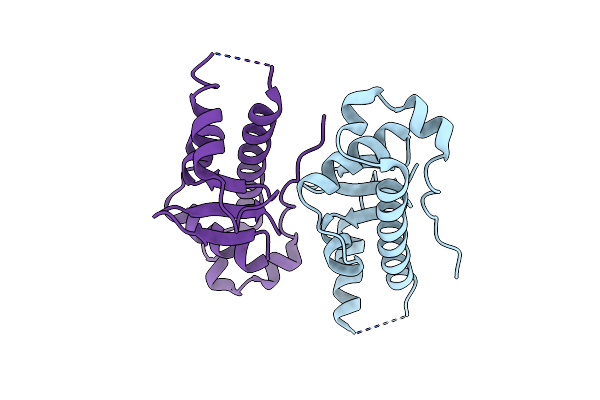 |
Structural Basis For The Microtubule Binding Of The Human Kinetochore Ska Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2014-01-22 Classification: CELL CYCLE |
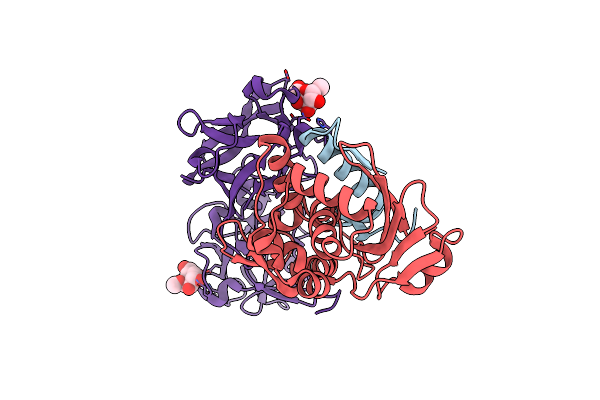 |
Crystal Structure Of Snake Gourd (Trichosanthes Anguina) Seed Lectin, A Three Chain Homologue Of Type Ii Rips
Organism: Trichosanthes anguina
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2013-08-07 Classification: SUGAR BINDING PROTEIN Ligands: AMG |
 |
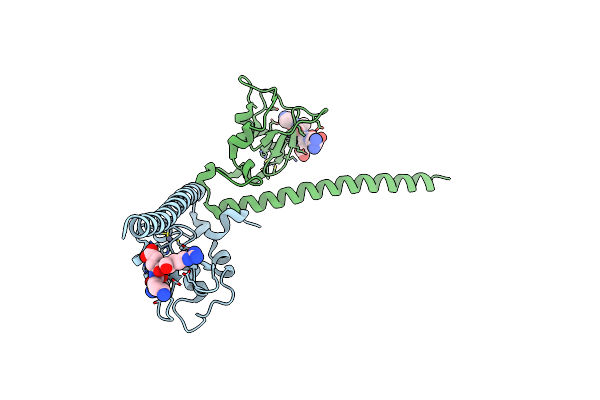
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.32 Å Release Date: 2012-05-23 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2011-11-09 Classification: CELL CYCLE Ligands: ZN |

