Search Count: 152
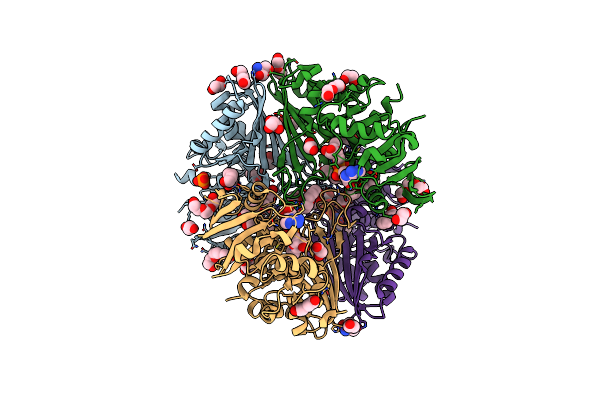 |
Structure Of The Complex Of Erythrose-4-Phosphate Dehydrogenase From Acinetobacter Baumannii With Nicotinamide Adenine Dinucleotide In The Presence Of Poly(Ethylene Glycol) At 2.20 A Resolution
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: NAD, PEG, PG4, EDO, PO4, PGE, TRS, 1PE, GOL |
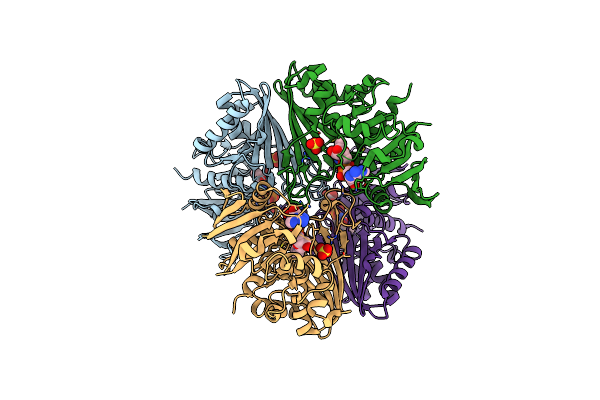 |
Structure Of The Complex Of Erythrose-4-Phosphate Dehydrogenase From Acinetobacter Baumannii With Nicotinamide Adenine Dinucleotide At 2.74 A Resolution.
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: NAD, SO4 |
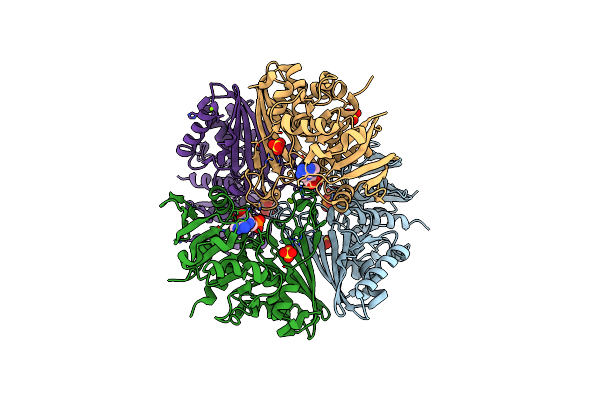 |
Crystal Structure Of The Complex Of Erythrose-4-Phosphate Dehydrogenase From Acinetobacter Baumannii With Adenosine Phosphate At 2.40 A Resolution.
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: AMP, SO4, MG |
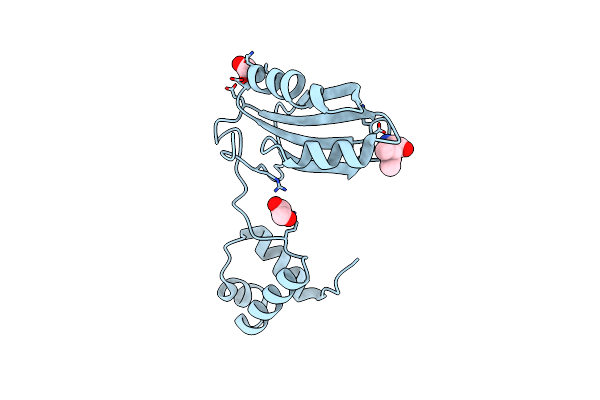 |
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-07-20 Classification: TRANSCRIPTION Ligands: ILE, EDO |
 |
Structure Of Hypothetical Protein Ttha1873 From Thermus Thermophilus With Potassium Mercuric Iodide
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-04-06 Classification: UNKNOWN FUNCTION Ligands: CA, 72I |
 |
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2022-03-09 Classification: UNKNOWN FUNCTION Ligands: CA |
 |
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2022-03-09 Classification: UNKNOWN FUNCTION Ligands: CA |
 |
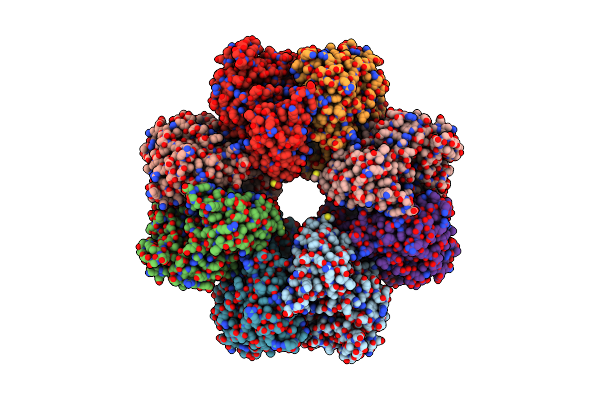
Mycobacterium Tuberculosis Enolase Mutant - E204A Complex With Phosphoenolpyruvate
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-02-16 Classification: HYDROLASE Ligands: PEP, PEG, EDO, ACT, MG |
 |
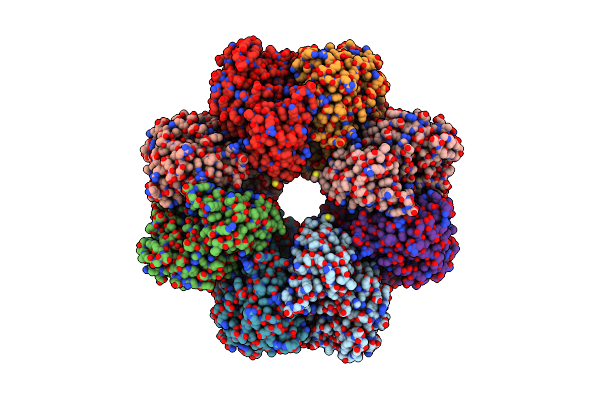
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2022-02-16 Classification: METAL BINDING PROTEIN |
 |
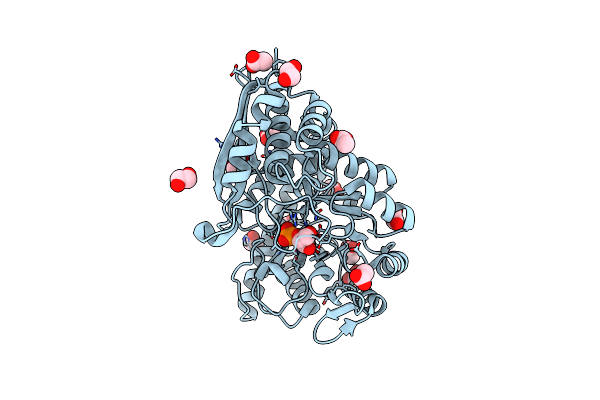
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2022-02-16 Classification: METAL BINDING PROTEIN Ligands: PEP, MG |
 |
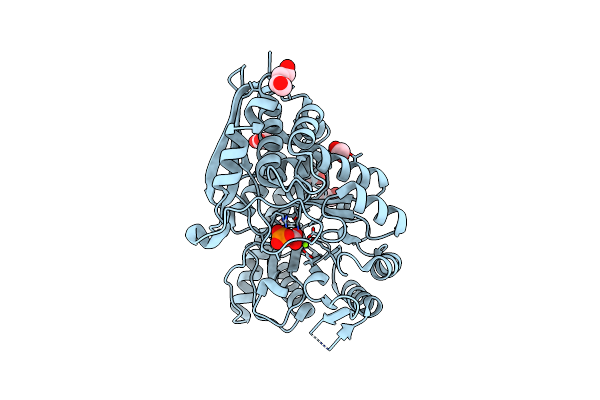
Mycobacterium Tuberculosis Enolase In Complex With Alternate 2-Phosphoglycerate
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-01-26 Classification: LYASE Ligands: EDO, ACT, MG, 2PG |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-12-01 Classification: LYASE Ligands: PEP, PEG, EDO, ACT, MG, CL |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2021-07-28 Classification: LYASE Ligands: MG, 2PG, GOL, ACT, PEG, CL |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2021-07-21 Classification: LYASE Ligands: MG, MPD |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-11-04 Classification: HYDROLASE Ligands: EDO, 2PG, ACT, MG |
 |
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-05-22 Classification: RECOMBINATION Ligands: ZN, CL |
 |
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2019-05-22 Classification: RECOMBINATION |
 |
Crystal Structure Of Thymidylate Kinase In Complex With Adp, Tdp And Tmp From Thermus Thermophilus Hb8
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2018-12-19 Classification: TRANSFERASE Ligands: ADP, MG, CL, TYD, TMP |
 |
Crystal Structure Of Thymidylate Kinase In Complex With Adp And Tdp From Thermus Thermophilus Hb8
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2018-12-19 Classification: TRANSFERASE Ligands: TYD, ADP, MG, CL |
 |
Crystal Structure Of Thymidylate Kinase In Complex With Adp And Tmp From Thermus Thermophilus Hb8
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2018-12-19 Classification: TRANSFERASE Ligands: ADP, CA, CL, TMP |

