Search Count: 24
 |
 |
 |
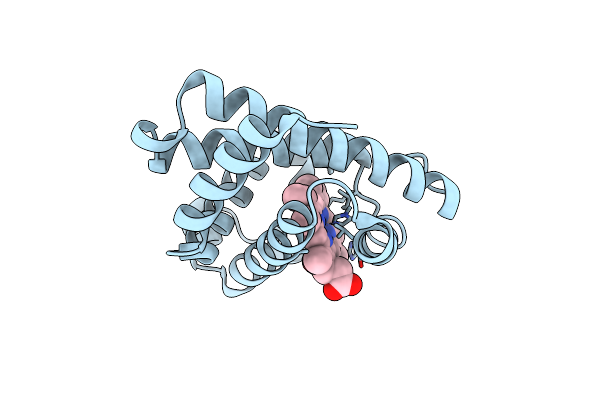 |
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-08-22 Classification: OXYGEN STORAGE Ligands: HEM |
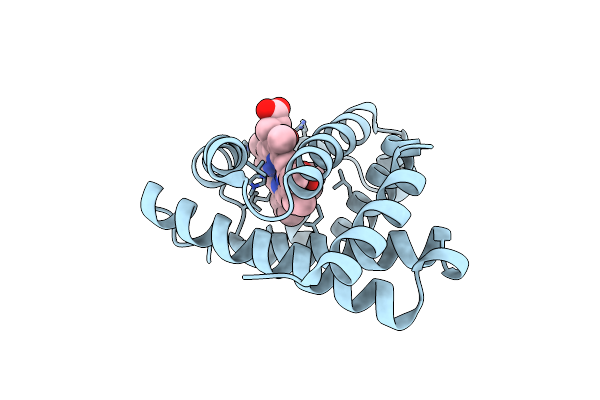 |
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-08-22 Classification: OXYGEN STORAGE Ligands: HEM, EEE |
 |
Structure Of Mb Nmh H64V, V68A Mutant Complex With Eda Incubated At Room Temperature For 5 Min
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-08-22 Classification: OXYGEN STORAGE Ligands: EEE, HEM |
 |
Structure Of Mb Nmh H64V, V68A Mutant Complex With Eda Incubated At Room Temperature For 20 Min
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-08-22 Classification: OXYGEN STORAGE Ligands: EEE, HEM |
 |
Heme-Carbene Complex In Myoglobin H64V/V68A Containing An N-Methylhistidine As The Proximal Ligand, 1.48 Angstrom Resolution
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2018-08-22 Classification: OXYGEN STORAGE Ligands: HEM, EEE |
 |
Heme-Carbene Complex In Myoglobin H64V/V68A Containing An N-Methylhistidine As The Proximal Ligand, 1.6 Angstrom Resolution
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-08-22 Classification: OXYGEN STORAGE Ligands: HEM, EEE |
 |
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-08-22 Classification: OXYGEN STORAGE Ligands: HEM, EDO |
 |
Organism: Homo sapiens, Synthetic construct
Method: SOLUTION NMR Release Date: 2018-02-21 Classification: RNA BINDING PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: SOLUTION NMR Release Date: 2018-02-21 Classification: RNA BINDING PROTEIN |
 |
Organism: Mycobacterium intracellulare 1956
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2017-11-01 Classification: MEMBRANE PROTEIN Ligands: ONM, MN, SO4 |
 |
X-Ray Structure Of A Bacterial Adenylyl Cyclase Soluble Domain, Solved At Cryogenic Temperature
Organism: Mycobacterium intracellulare 1956
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-11-01 Classification: MEMBRANE PROTEIN Ligands: ONM, MN, SO4 |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2016-12-21 Classification: LIPID BINDING PROTEIN |
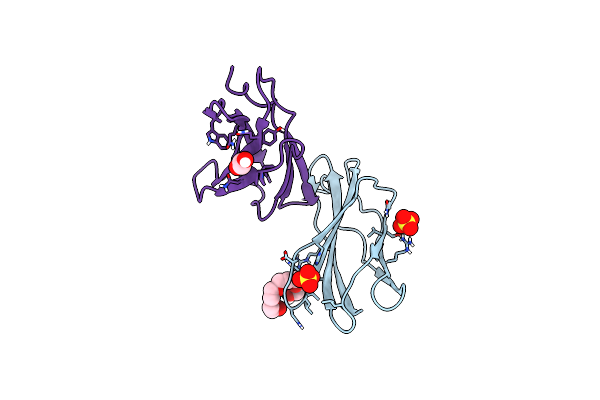 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2015-02-11 Classification: CELL ADHESION Ligands: SO4, 1PE, EDO |
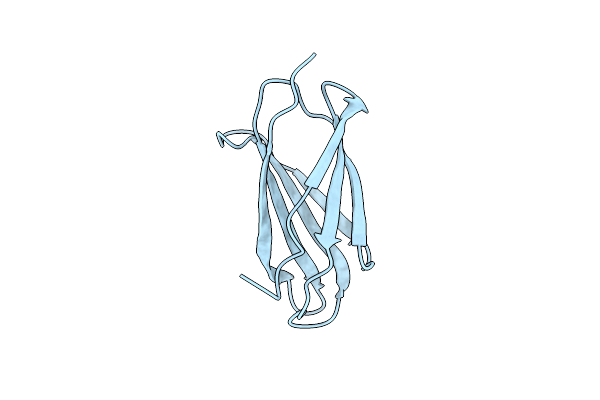 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-02-11 Classification: CELL ADHESION |
 |
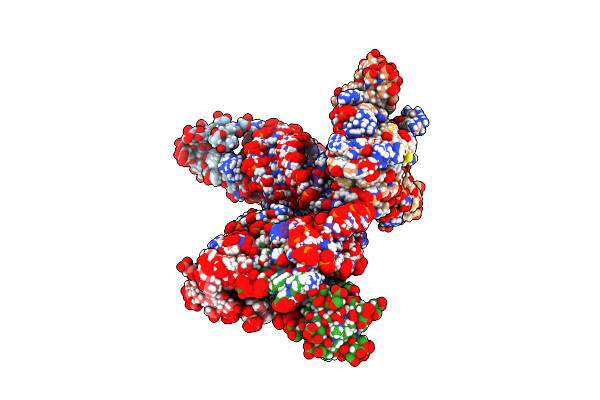
Structural Basis Of The Non-Coding Rna Rsmz Acting As Protein Sponge: Conformer L Of Rsmz(1-72)/Rsme(Dimer) 1To3 Complex
Organism: Pseudomonas protegens pf-5
Method: SOLUTION NMR Release Date: 2014-05-21 Classification: RNA BINDING PROTEIN/RNA |
 |
Structural Basis Of The Non-Coding Rna Rsmz Acting As Protein Sponge: Conformer R Of Rsmz(1-72)/Rsme(Dimer) 1To3 Complex
Organism: Pseudomonas protegens pf-5
Method: SOLUTION NMR Release Date: 2014-05-21 Classification: RNA BINDING PROTEIN/RNA |
 |
Crystal Structure Of The Small Terminase Oligomerization Core Domain From A Spp1-Like Bacteriophage (Crystal Form 1)
Organism: Bacillus phage sf6
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-12-28 Classification: DNA BINDING PROTEIN |

