Search Count: 28
 |
Organism: Coxiella burnetii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-08-13 Classification: CYTOSOLIC PROTEIN Ligands: EPE, EDO, PEG, CL |
 |
Crystal Structure Of The Coxiella Burnetii E110Q Mutant 2-Methylisocitrate Lyase
Organism: Coxiella burnetii
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2025-08-13 Classification: CYTOSOLIC PROTEIN Ligands: EDO, CL |
 |
Crystal Structure Of The Coxiella Burnetii 2-Methylisocitrate Lyase Bound To Substrate 2-Mic
Organism: Coxiella burnetii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-08-13 Classification: CYTOSOLIC PROTEIN Ligands: MIC, EOH, EDO, MG, CL |
 |
Crystal Structure Of The Coxiella Burnetii R152Q Mutant 2-Methylisocitrate Lyase
Organism: Coxiella burnetii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-08-13 Classification: CYTOSOLIC PROTEIN Ligands: CIT, EDO, CL |
 |
Crystal Structure Of The Coxiella Burnetii 2-Methylisocitrate Lyase Bound To Inhibitor Isocitric Acid
Organism: Coxiella burnetii
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2025-08-13 Classification: CYTOSOLIC PROTEIN Ligands: ICT, EDO, MG, CL, PEG, PGE |
 |
Crystal Structure Of The Coxiella Burnetii 2-Methylisocitrate Lyase Bound To Products Succinic And Pyruvic Acid
Organism: Coxiella burnetii
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2025-08-13 Classification: CYTOSOLIC PROTEIN Ligands: SIN, PYR, EDO, MG, CL, DMS, PGE, PEG |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2024-09-11 Classification: HYDROLASE Ligands: GOL, SO4, MN, MG |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-06-14 Classification: RNA BINDING PROTEIN Ligands: NN9, CIT |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-06-14 Classification: RNA BINDING PROTEIN Ligands: NMV, CIT |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-06-14 Classification: RNA BINDING PROTEIN Ligands: NLL, CIT |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-06-14 Classification: RNA BINDING PROTEIN Ligands: CIT, NL1 |
 |
Cryo-Em Structure Of Sars-Cov-2 Spike In Complex With Antibodies B1-182.1 And A19-61.1
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: VIRAL PROTEIN/Immune System Ligands: NAG |
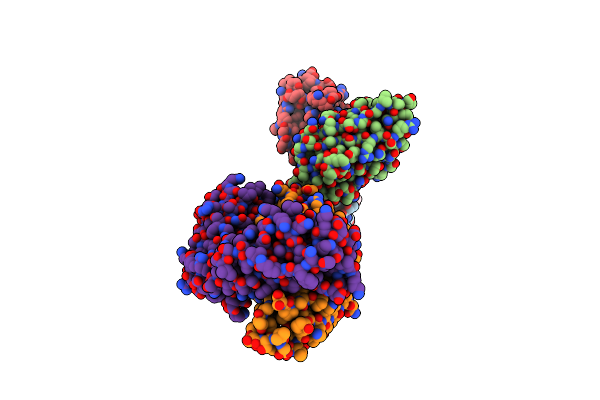 |
Locally Refined Region Of Sars-Cov-2 Spike In Complex With Antibodies B1-182.1 And A19-61.1
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: VIRAL PROTEIN/Immune System |
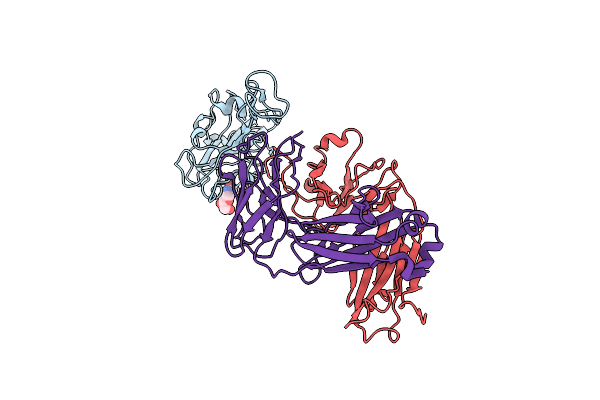 |
Locally Refined Region Of Sars-Cov-2 Spike In Complex With Antibody A19-46.1
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: VIRAL PROTEIN/Immune System Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 Omicron Spike In Complex With Antibody A19-46.1
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: VIRAL PROTEIN/Immune System Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 Omicron Spike In Complex With Antibodies A19-46.1 And B1-182.1
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: VIRAL PROTEIN/Immune System Ligands: NAG |
 |
Local Refinement Of Cryo-Em Structure Of The Interface Of The Omicron Rbd In Complex With Antibodies B-182.1 And A19-46.1
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-03-30 Classification: Vral Protein/IMMUNE SYSTEM Ligands: NAG |
 |
Solution Structure Of The Razul Domain From 26S Proteasome Subunit Hrpn10/S5A Complexed With The Azul Domain From E3 Ligase E6Ap/Ube3A
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2020-03-18 Classification: STRUCTURAL PROTEIN Ligands: ZN |
 |
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2019-09-11 Classification: OXIDOREDUCTASE |
 |
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2019-08-14 Classification: LYASE |

