Search Count: 48
 |
Protein 2A From Theiler'S Murine Encephalomyelitis Virus (Tmev) Bound To Rna Pseudoknot
Organism: Theiler's encephalomyelitis virus
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: RNA |
 |
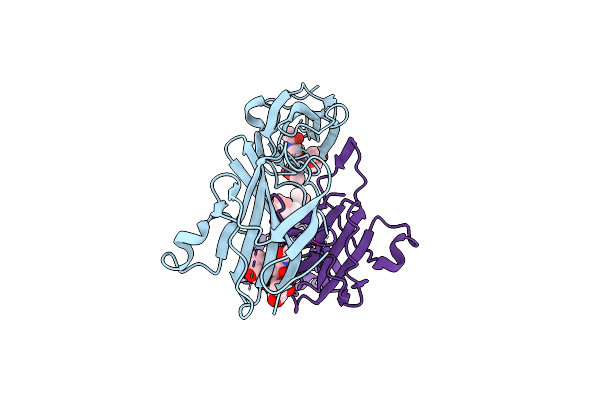
Structure Of The N-Terminal Didomain D1_D2 Of The Thrombospondin Type-1 Domain-Containing 7A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-05-15 Classification: CELL ADHESION |
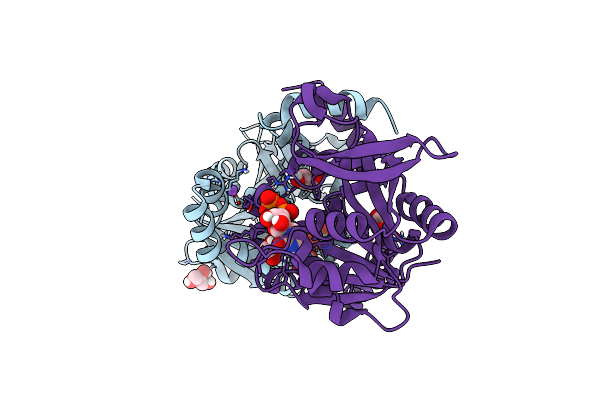 |
Crystal Structure Of Udp-Glucose Pyrophosphorylase From Thermocrispum Agreste Dsm 44070 In Complex With Udp
Organism: Thermocrispum agreste dsm 44070
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-01-17 Classification: TRANSFERASE Ligands: UDP, GOL, NA, EDO |
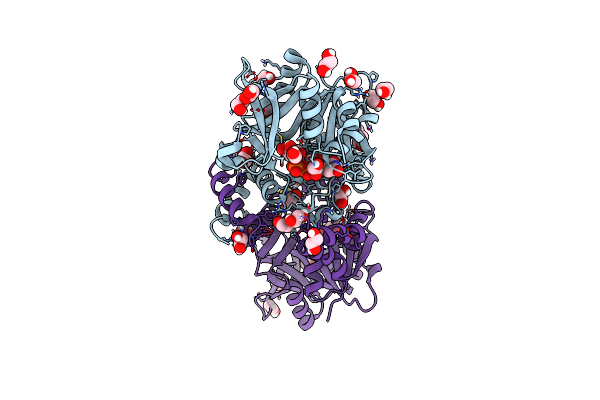 |
Crystal Structure Of Udp-Glucose Pyrophosphorylase From Thermocrispum Agreste Dsm 44070 In Complex With Udp-Glucose
Organism: Thermocrispum agreste dsm 44070
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-01-10 Classification: TRANSFERASE Ligands: UPG, EDO, PEG, SCN, MG |
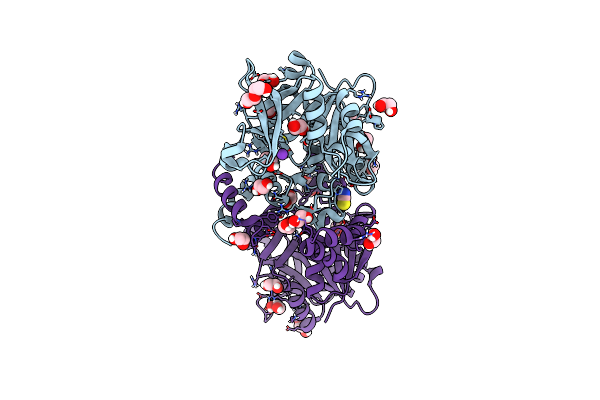 |
Crystal Structure Of Udp-Glucose Pyrophosphorylase From Thermocrispum Agreste Dsm 44070
Organism: Thermocrispum agreste dsm 44070
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-12-27 Classification: TRANSFERASE Ligands: PEG, EDO, SCN, K |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2023-03-15 Classification: CELL ADHESION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2023-03-15 Classification: CELL ADHESION Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-03-15 Classification: CELL ADHESION Ligands: NAG, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2023-03-08 Classification: CELL ADHESION Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-03-08 Classification: CELL ADHESION Ligands: NAG |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2023-02-08 Classification: LIGASE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2023-02-08 Classification: LIGASE Ligands: SO4 |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-02-08 Classification: LIGASE |
 |
Crystal Structure Of Holo-Swhpa-Mg (Hydroxy Ketone Aldolase) From Sphingomonas Wittichii Rw1 In Complex With Hydroxypyruvate And D-Glyceraldehyde
Organism: Rhizorhabdus wittichii rw1
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-11-23 Classification: LYASE Ligands: 3PY, 3GR, PEG, MG, BR, K |
 |
Crystal Structure Of S116A Mutant Of Hydroxy Ketone Aldolase (Swhka) From Sphingomonas Wittichii Rw1 In Complex With Hydroxypyruvate
Organism: Sphingomonas wittichii (strain rw1 / dsm 6014 / jcm 10273)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-11-16 Classification: LYASE Ligands: 3PY, MG |
 |
Crystal Structure Of Apo-Swhka (Hydroxy Ketone Aldolase) From Sphingomonas Wittichii Rw1
Organism: Sphingomonas wittichii (strain rw1 / dsm 6014 / jcm 10273)
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2022-11-16 Classification: LYASE Ligands: PEG, BR, K |
 |
Crystal Structure Of Holo-H44A Mutant Of Hydroxy Ketone Aldolase (Swhka) From Sphingomonas Wittichii Rw1, In Complex With Hydroxypyruvate
Organism: Sphingomonas wittichii (strain rw1 / dsm 6014 / jcm 10273)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-11-16 Classification: LYASE Ligands: MG, BR, K, 3PY |
 |
Crystal Structure Of Holo-Swhpa-Mn (Hydroxyketoacid Aldolase) From Sphingomonas Wittichii Rw1
Organism: Rhizorhabdus wittichii rw1
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-10-26 Classification: LYASE Ligands: MN, K, BR |
 |
Crystal Structure Of Holo-F210W Mutant Of Hydroxy Ketone Aldolase (Swhka)From Sphingomonas Wittichii Rw1
Organism: Sphingomonas wittichii (strain rw1 / dsm 6014 / jcm 10273)
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-10-26 Classification: LYASE Ligands: PEG, MG, BR, K |
 |
Crystal Structure Of Holo-F210W Mutant Of Hydroxy Ketone Aldolase (Swhka) From Sphingomonas Wittichii Rw1 In Complex With Hydroxypyruvate
Organism: Sphingomonas wittichii (strain rw1 / dsm 6014 / jcm 10273)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-10-26 Classification: LYASE Ligands: 3PY, PEG, MG, K, BR |

