Search Count: 198
 |
Organism: Xenopus laevis, Bos taurus
Method: ELECTRON MICROSCOPY Resolution:2.89 Å Release Date: 2025-11-05 Classification: CELL CYCLE Ligands: G2P, MG, GTP |
 |
An Alpha-L-Arabinofuranosidase (Atabf43C) From Acetivibrio Thermocellus Dsm1313
Organism: Acetivibrio thermocellus dsm 1313
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2025-08-20 Classification: HYDROLASE Ligands: GOL, MG |
 |
An Alpha-L-Arabinofuranosidase (Atabf43C) From Acetivibrio Thermocellus Dsm1313 Bound To Arabinofuranose
Organism: Acetivibrio thermocellus dsm 1313
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-08-20 Classification: HYDROLASE Ligands: AHR, GOL, MG |
 |
Cbm42 Domain Of Alpha-L-Arabinofuranosidase (Atabf43C) From Acetivibrio Thermocellus Dsm1313
Organism: Acetivibrio thermocellus dsm 1313
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-08-20 Classification: HYDROLASE Ligands: GOL |
 |
The Gh43 Domain Of An Alpha-L-Arabinofuranosidase (Atabf43C_Gh43) From Acetivibrio Thermocellus Dsm1313
Organism: Acetivibrio thermocellus dsm 1313
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2025-08-20 Classification: HYDROLASE Ligands: AHR, SO4 |
 |
Organism: Tenacibaculum discolor
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-08-13 Classification: TRANSFERASE Ligands: A1BIH, PEG |
 |
Acla From Tenacibaculum Discolor In Complex With The C8-N-Acyl Cyclolysine Reaction Product (C8-Acl)
Organism: Tenacibaculum discolor
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2025-08-13 Classification: TRANSFERASE Ligands: A1CA3, MAE, PEG |
 |
Acla From Tenacibaculum Discolor In Complex With Intermediate Formed By C8-Lysine Attack On Adp
Organism: Tenacibaculum discolor
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2025-08-13 Classification: TRANSFERASE Ligands: AMP, A1BIH, PEG |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2023-10-04 Classification: TRANSPORT PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:0.96 Å Release Date: 2023-06-28 Classification: VIRAL PROTEIN Ligands: GOL, KZ0 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-06-28 Classification: VIRAL PROTEIN Ligands: 29N |
 |
Crystal Structure Of The Snare Use1 Bound To Dsl1 Complex Subunits Sec39 And Dsl1, Revised Use1 Structure
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Resolution:5.73 Å Release Date: 2023-03-01 Classification: TRANSPORT PROTEIN |
 |
Organism: Amycolatopsis keratiniphila
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2022-12-07 Classification: ANTIBIOTIC Ligands: MAN, YBJ, FMT, CL |
 |
Organism: Amycolatopsis keratiniphila
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2022-12-07 Classification: ANTIBIOTIC Ligands: MAN, YBJ, FMT |
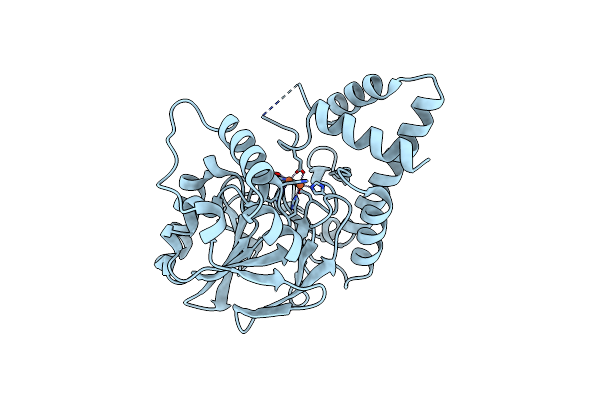 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2022-08-31 Classification: HYDROLASE Ligands: FE |
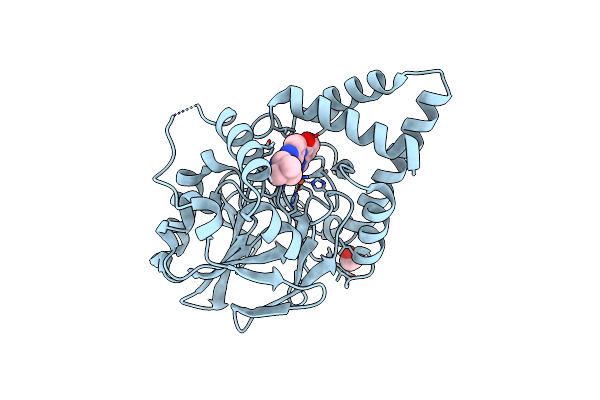 |
Structure Of Pqs Response Protein Pqse In Complex With N-(4-(3-Neopentylureido)Phenyl)-1H-Indazole-7-Carboxamide
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-08-31 Classification: HYDROLASE/INHIBITOR Ligands: FE, KYX, EDO |
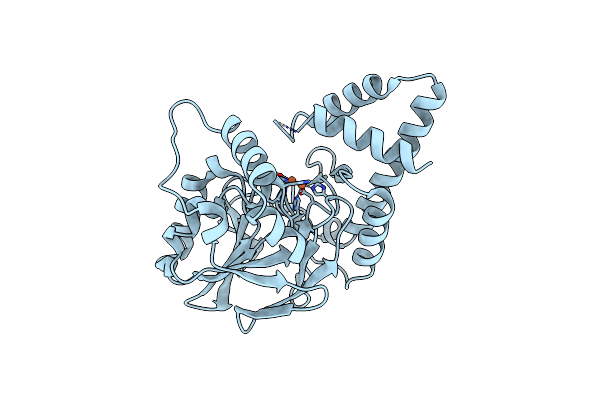 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2022-08-31 Classification: HYDROLASE Ligands: FE |
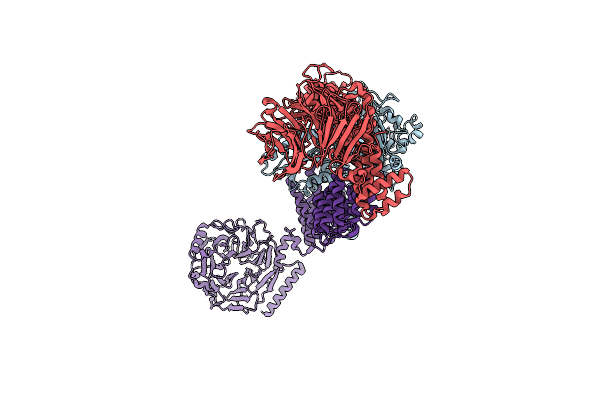 |
Cryo-Em Structure Of A Hops Core Complex Containing Vps33, Vps16, And Vps18
Organism: Chaetomium thermophilum
Method: ELECTRON MICROSCOPY Resolution:5.10 Å Release Date: 2022-08-31 Classification: PROTEIN TRANSPORT |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2022-05-25 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-05-04 Classification: TRANSFERASE Ligands: NAD |

