Search Count: 26
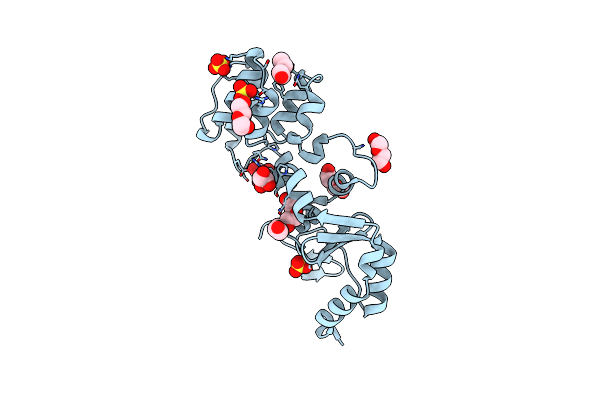 |
The Structure Of A Sensor Domain Of A Histidine Kinase (Vxra) From Vibrio Cholerae O1 Biovar Eltor Str. N16961, N239 Deletion Mutant
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2021-01-27 Classification: SIGNALING PROTEIN Ligands: GOL, PEG, PG4, SO4 |
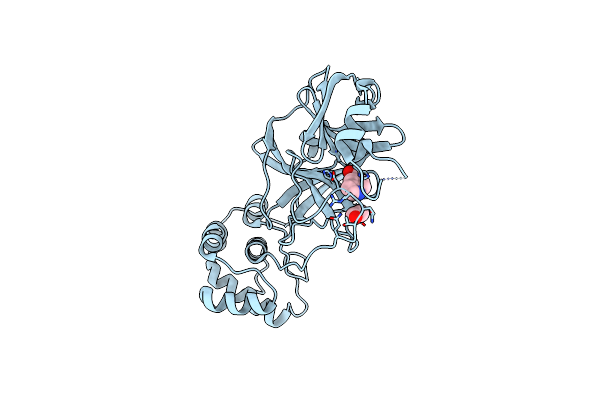 |
The Crystal Structure Of Sars-Cov-2 Main Protease With The Formation Of Cys145-1H-Indole-5-Carboxylate
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2020-12-16 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: XC4, EDO |
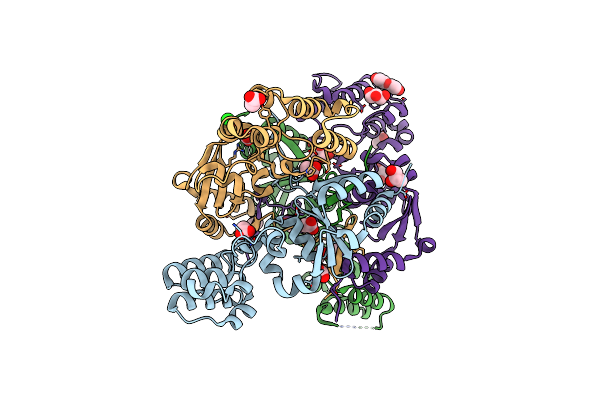 |
The Structure Of A Sensor Domain Of A Histidine Kinase (Vxra) From Vibrio Cholerae O1 Biovar Eltor Str. N16961, 2Nd Form
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-10-14 Classification: SIGNALING PROTEIN Ligands: GOL, ACT, CL, PEG, SRT |
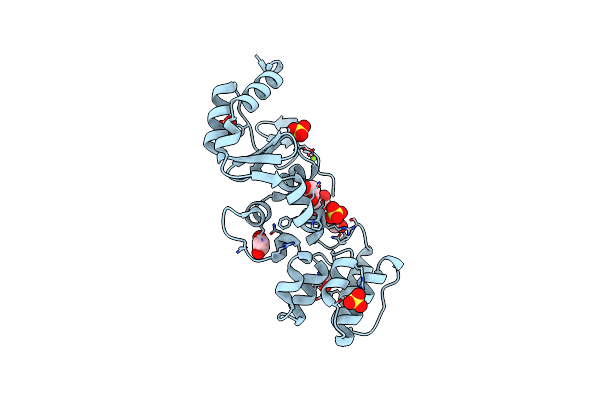 |
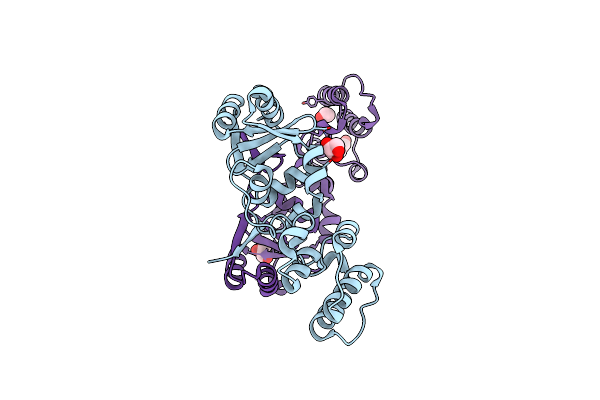
The Structure Of A Sensor Domain Of A Histidine Kinase (Vxra) From Vibrio Cholerae O1 Biovar Eltor Str. N16961, N239-T240 Deletion Mutant
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-10-14 Classification: SIGNALING PROTEIN Ligands: EDO, SO4, MG |
 |
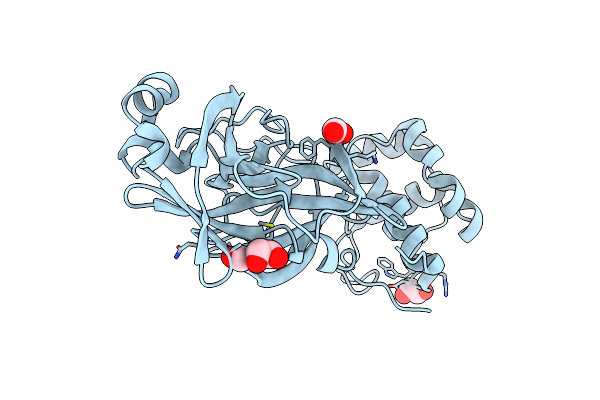
The Structure Of A Sensor Domain Of A Histidine Kinase (Vxra) From Vibrio Cholerae O1 Biovar Eltor Str. N16961, D238-T240 Deletion Mutant
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2020-10-14 Classification: SIGNALING PROTEIN Ligands: GOL, EDO |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-07-29 Classification: HYDROLASE Ligands: EDO, FMT |
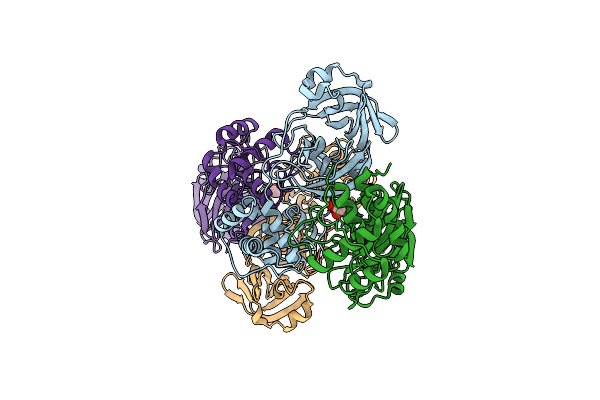 |
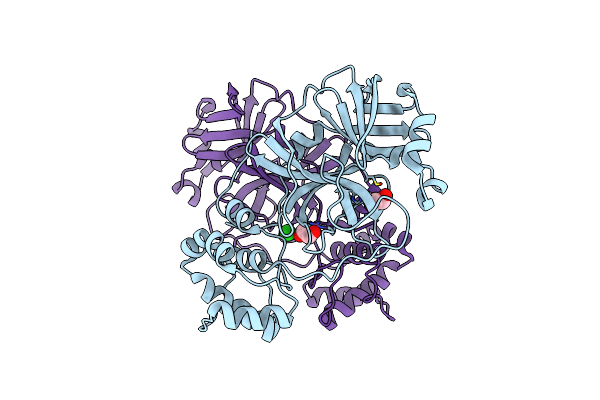
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-07-15 Classification: HYDROLASE Ligands: EDO |
 |
The Crystal Structure Of 3Cl Mainpro Of Sars-Cov-2 With Oxidized Cys145 (Sulfenic Acid Cysteine).
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: EDO, CL |
 |
The 1.28A Crystal Structure Of 3Cl Mainpro Of Sars-Cov-2 With Oxidized C145 (Sulfinic Acid Cysteine)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: EDO, FMT, ACT |
 |
The Structure Of Condensation And Adenylation Domains Of Teixobactin-Producing Nonribosomal Peptide Synthetase Txo1 Serine Module In Complex With Mg And Amp
Organism: Eleftheria terrae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-06-12 Classification: BIOSYNTHETIC PROTEIN Ligands: AMP, MG, SO4, CL, ACT, GOL, FMT |
 |
The Structure Of Condensation And Adenylation Domains Of Teixobactin-Producing Nonribosomal Peptide Synthetase Txo1 Serine Module In Complex With Mg
Organism: Eleftheria terrae
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-06-05 Classification: BIOSYNTHETIC PROTEIN Ligands: MG, MES, SO4 |
 |
The Structure Of Condensation And Adenylation Domains Of Teixobactin-Producing Nonribosomal Peptide Synthetase Txo1 Serine Module
Organism: Eleftheria terrae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-05-29 Classification: BIOSYNTHETIC PROTEIN Ligands: FMT, SO4, EPE, MES, GOL |
 |
The Structure Of Condensation And Adenylation Domains Of Teixobactin-Producing Nonribosomal Peptide Synthetase Txo1 Serine Module In Complex With Amp
Organism: Eleftheria terrae
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2019-05-29 Classification: BIOSYNTHETIC PROTEIN Ligands: AMP, SO4, GOL |
 |
The Structure Of Condensation And Adenylation Domains Of Teixobactin-Producing Nonribosomal Peptide Synthetase Txo2 Serine Module
Organism: Eleftheria terrae
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2019-05-29 Classification: BIOSYNTHETIC PROTEIN Ligands: FMT, FLC, ACT, CL, GOL, SO4 |
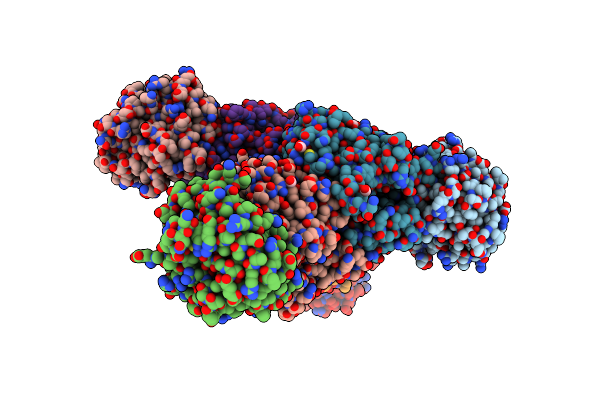 |
Crystal Structure Of Tryptophan Synthase From M. Tuberculosis - Ligand-Free Form
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2017-05-31 Classification: LYASE Ligands: MLI, FMT, K |
 |
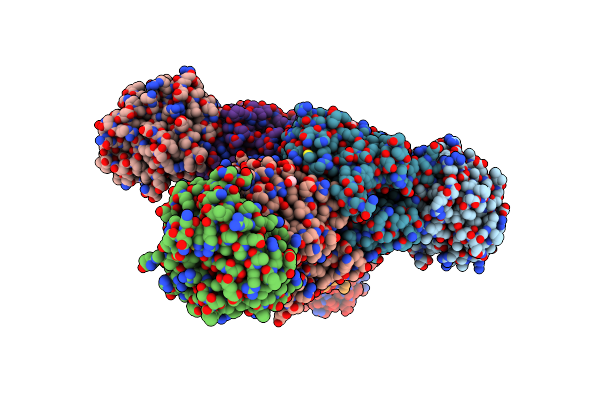
Crystal Structure Of Tryptophan Synthase From M. Tuberculosis - Aminoacrylate-Bound Form
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-05-31 Classification: LYASE Ligands: MLI, FMT, P1T, CS |
 |
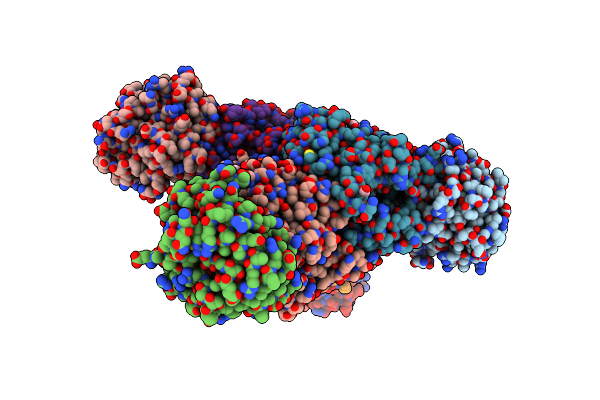
Crystal Structure Of Tryptophan Synthase From M. Tuberculosis - Ligand-Free Form, Trpa-G66V Mutant
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2017-05-31 Classification: LYASE Ligands: MLI, FMT |
 |
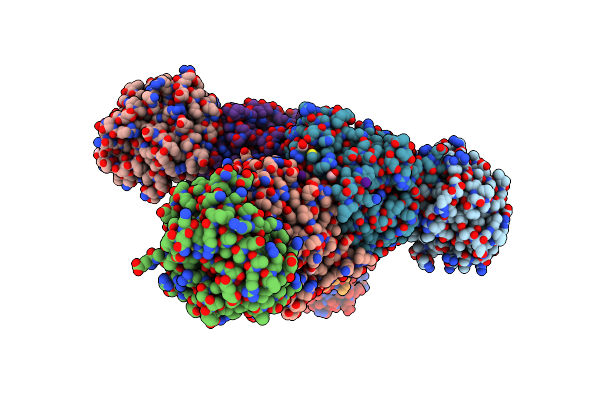
Crystal Structure Of Tryptophan Synthase From M. Tuberculosis - Brd4592-Bound Form
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2017-05-31 Classification: LYASE Ligands: MLI, FMT, 79V |
 |
Crystal Structure Of Tryptophan Synthase From M. Tuberculosis - Aminoacrylate And Brd4592-Bound Form
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-05-31 Classification: LYASE Ligands: MLI, FMT, P1T, CS, 79V |
 |
The 2.4A Crystal Structure Of Ompa Domain Of Ompa From Salmonella Enterica Subsp. Enterica Serovar Typhimurium Str. 14028S
Organism: Salmonella typhimurium (strain 14028s / sgsc 2262)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-04-19 Classification: MEMBRANE PROTEIN Ligands: SO4 |

