Search Count: 48
 |
Structure Of Full-Length Human Protein Kinase C Beta 2 (Pkcbii) In The Inactive Conformation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: SIGNALING PROTEIN Ligands: ZN, ANP, MG |
 |
Structure Of Full-Length Human Protein Kinase C Beta 1 (Pkcbi) In The Active And Inactive Conformation Soaked In Manganese Chloride
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: SIGNALING PROTEIN,TRANSFERASE Ligands: GOL, ZN, ANP, MN, CA |
 |
Structure Of Full-Length Human Protein Kinase C Beta 1 (Pkcbi) In The Active Conformation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: SIGNALING PROTEIN,TRANSFERASE Ligands: GOL, ZN |
 |
Structure Of Full-Length Human Protein Kinase C Beta 1 (Pkcbi) In The Active And Inactive Conformation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: SIGNALING PROTEIN, TRANSFERASE Ligands: ZN, ADN |
 |
Structural Basis Of Bak Sequestration By Mcl-1 And Consequences For Apoptosis Initiation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2025-06-04 Classification: APOPTOSIS |
 |
Structural Basis Of Bak Sequestration By Mcl-1 And Consequences For Apoptosis Initiation
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-06-04 Classification: APOPTOSIS |
 |
Structural Basis Of Bak Sequestration By Mcl-1 And Consequences For Apoptosis Initiation
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: APOPTOSIS |
 |
Structural Basis Of Bak Sequestration By Mcl-1 And Consequences For Apoptosis Initiation
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-06-04 Classification: APOPTOSIS |
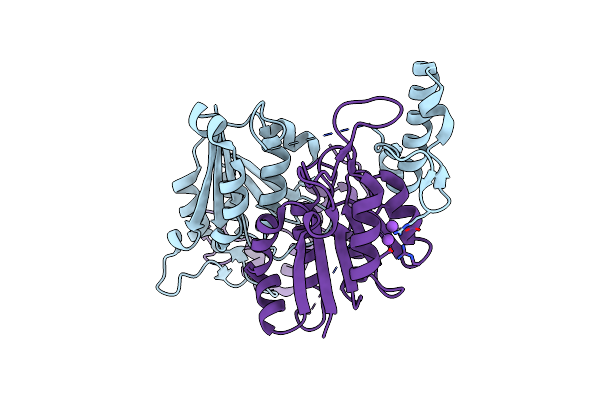 |
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2023-01-18 Classification: TRANSFERASE |
 |
Mycobacterium Abscessus Trna Methyltransferase In Complex With S-Adenosyl-L-Homocysteine
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: SAH |
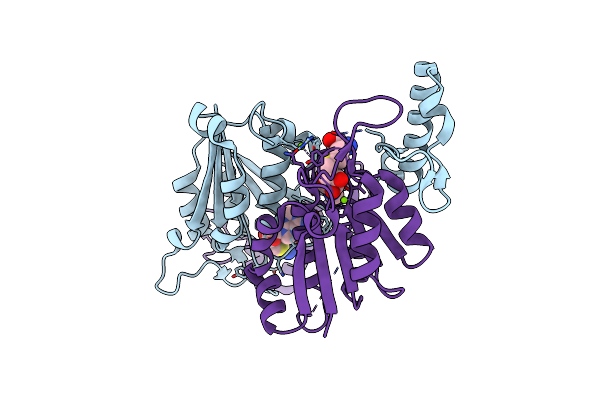 |
Mycobacterium Abscessus Trna Methyltransferase In Complex With S-Adenosyl-L-Homocysteine And Magnesium
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: SAH, MG |
 |
The Crystal Structure Of The I38T Mutant Pa Endonuclease (2009/H1N1/California) In Complex With Sj000988558
Organism: Influenza a virus, Influenza a virus (a/luxembourg/43/2009(h1n1))
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2021-11-10 Classification: VIRAL PROTEIN Ligands: MN, WTD, QQ4, SO4 |
 |
The Crystal Structure Of The I38T Mutant Pa Endonuclease (2009/H1N1/California) In Complex With Sj000988528
Organism: Influenza a virus, Influenza a virus (a/luxembourg/43/2009(h1n1))
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2021-11-10 Classification: VIRAL PROTEIN, Hydrolase Ligands: MN, WTG, QQ4, SO4 |
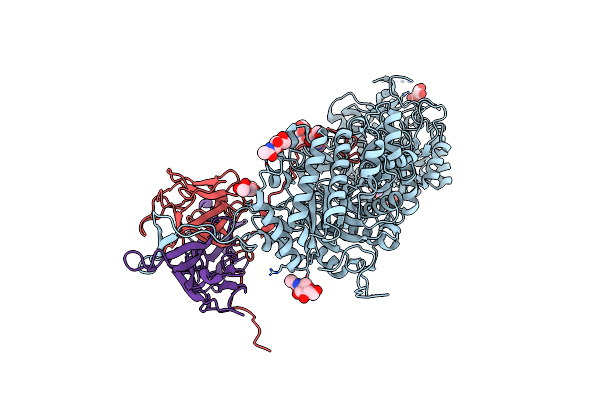 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-01-24 Classification: HYDROLASE/PROTEIN BINDING Ligands: NAG, ZN |
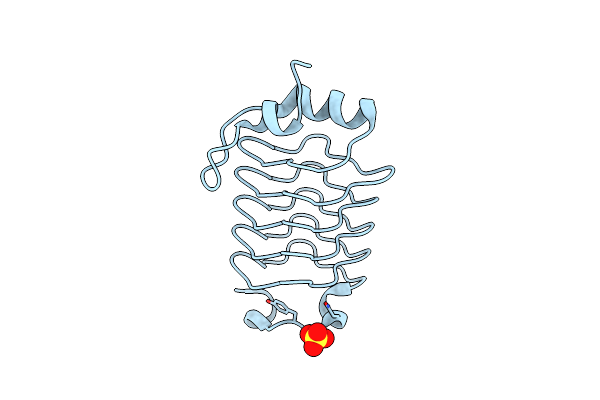 |
The 1.7 Angstrom Resolution Crystal Structure Of At2G44920, A Pentapeptide Repeat Protein From Arabidopsis Thaliana Thylakoid Lumen.
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-06-29 Classification: UNKNOWN FUNCTION Ligands: SO4 |
 |
Crystal Structure Of Sars-Cov Main Protease Triple Mutant Sti/A In Space Group P21
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2009-09-01 Classification: HYDROLASE |
 |
Crystal Structure Of Sars-Cov Main Protease Triple Mutant Sti/A In Space Group C2
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2009-09-01 Classification: HYDROLASE |
 |
Crystal Structure Of Sars-Cov Main Protease Quadruple Mutant Stif/A With One Molecule In One Asymmetric Unit
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2009-09-01 Classification: HYDROLASE |
 |
Crystal Structure Of Sars-Cov Main Protease Quadruple Mutant Stif/A With Two Molecules In One Asymmetric Unit
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2009-09-01 Classification: HYDROLASE |
 |
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2009-08-25 Classification: HYDROLASE |

