Search Count: 14
 |
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2023-01-11 Classification: DNA BINDING PROTEIN Ligands: ZN, MG |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-08-11 Classification: RNA BINDING PROTEIN Ligands: 1PE, TRS |
 |
Organism: Saccharomyces cerevisiae, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2016-06-15 Classification: TRANSLATION |
 |
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2016-01-13 Classification: RNA BINDING PROTEIN Ligands: HOH |
 |
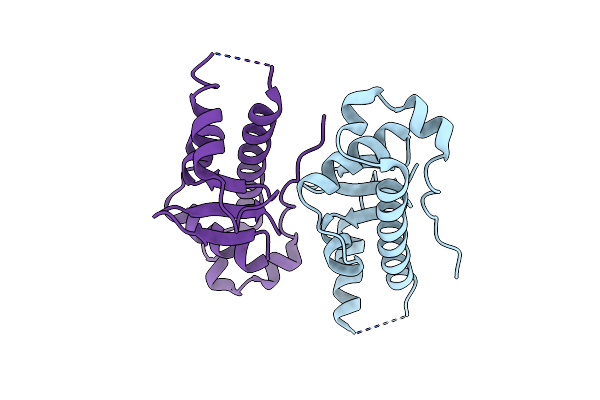
Structural Basis For The Microtubule Binding Of The Human Kinetochore Ska Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2014-01-22 Classification: CELL CYCLE |
 |
Structural Basis For The Microtubule Binding Of The Human Kinetochore Ska Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2014-01-22 Classification: CELL CYCLE |
 |
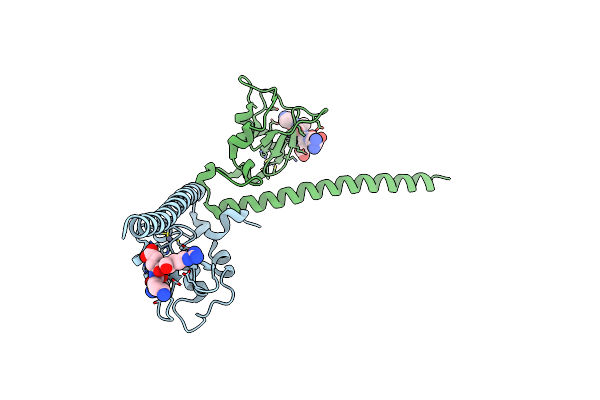
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.32 Å Release Date: 2012-05-23 Classification: CELL CYCLE |
 |
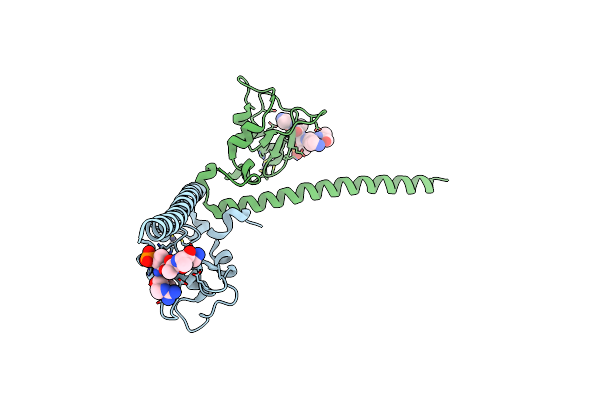
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2011-11-09 Classification: CELL CYCLE Ligands: ZN |
 |
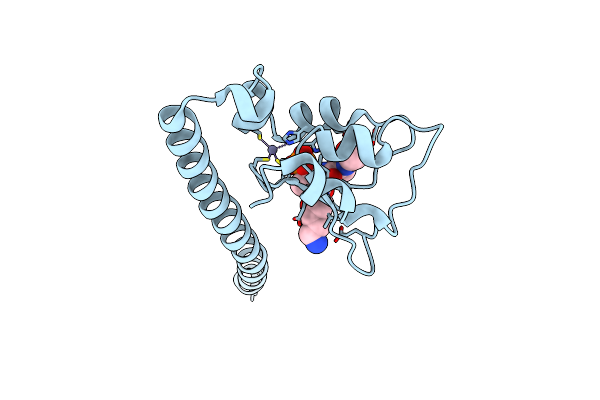
Crystal Structure Of Survivin Bound To The Phosphorylated N-Terminal Tail Of Histone H3
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2011-11-09 Classification: CELL CYCLE Ligands: ZN |
 |
Crystal Structure Of Survivin Bound To The Phosphorylated N-Terminal Tail Of Histone H3
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2011-11-09 Classification: CELL CYCLE Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2011-03-30 Classification: HYDROLASE/RNA Ligands: ZN, ADP, ALF, MG, 1PE |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2011-03-30 Classification: HYDROLASE/RNA Ligands: 1PE, ADP, ALF, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2011-03-30 Classification: HYDROLASE Ligands: GOL, MLI |
 |
Crystal Structure Of The Core Mago-Y14-Eif4Aiii-Barentsz-Upf3B Assembly Shows How The Ejc Is Bridged To The Nmd Machinery
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2010-05-12 Classification: HYDROLASE Ligands: MG, ANP |

