Search Count: 34
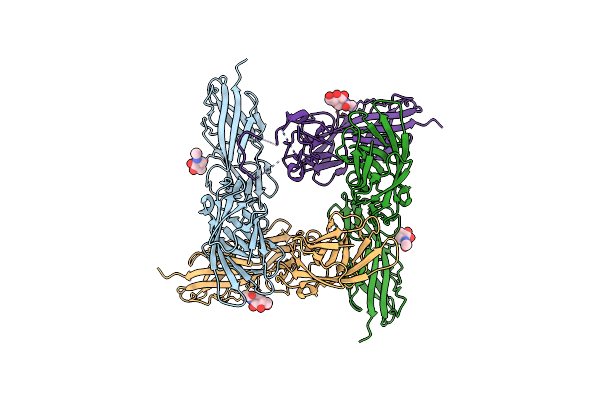 |
Structure-Based Engineering Of A Highly Immunogenic, Conformationally Stabilized Fimh Antigen For A Urinary Tract Infection Vaccine
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-02-05 Classification: CELL ADHESION Ligands: NAG |
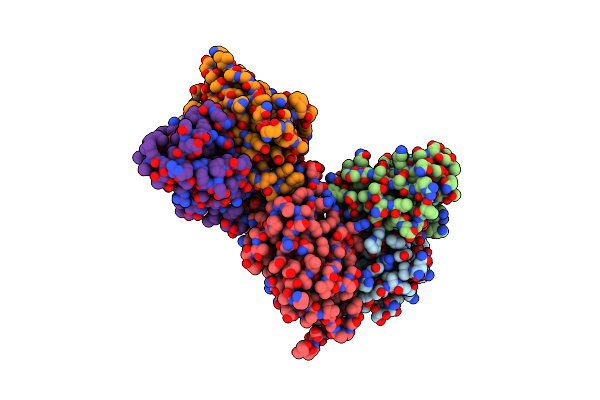 |
Cryo-Em Structure Of E. Coli Fimh Lectin Domain Bound To Fabs 440-2 And 454-3
Organism: Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: CELL ADHESION,SUGAR BINDING PROTEIN |
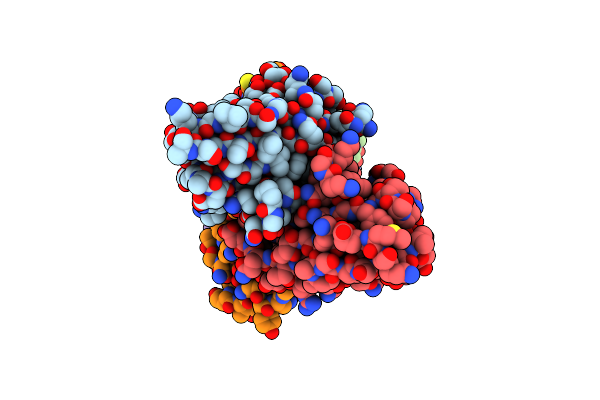 |
Cryo-Em Structure Of E. Coli Fimh Lectin Domain Bound To Fabs 329-2 And 454-3
Organism: Mus musculus, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: CELL ADHESION/IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2021-01-13 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO, O4B, VOJ, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2021-01-13 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO, O4B, VOM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2021-01-13 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO, O4B, VOP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2021-01-13 Classification: OXYGEN TRANSPORT Ligands: HEM, OXY, O4B, VUD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2020-09-23 Classification: TRANSFERASE Ligands: RYS, SO4, CIT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-09-23 Classification: TRANSFERASE Ligands: RZA, SO4 |
 |
Structure Of Khk In Complex With Compound 4 (6-[(1~{S},5~{R})-6-(Hydroxymethyl)-3-Azabicyclo[3.1.0]Hexan-3-Yl]-2-[(2~{S},3~{R})-2-Methyl-3-Oxidanyl-Azetidin-1-Yl]-4-(Trifluoromethyl)Pyridine-3-Carbonitrile)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2020-09-23 Classification: TRANSFERASE Ligands: S6J, SO4 |
 |
Structure Of Khk In Complex With Compound 6 (2-[(1~{R},5~{S})-3-[5-Cyano-6-[(2~{S},3~{R})-2-Methyl-3-Oxidanyl-Azetidin-1-Yl]-4-(Trifluoromethyl)Pyridin-2-Yl]-3-Azabicyclo[3.1.0]Hexan-6-Yl]Ethanoic Acid)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2020-09-23 Classification: TRANSFERASE Ligands: S6S, SO4 |
 |
Structure Of Khk In Complex With Compound 8 (2-[(1~{S},5~{R})-3-[2-[(2~{S})-2-Methylazetidin-1-Yl]-6-(Trifluoromethyl)Pyrimidin-4-Yl]-3-Azabicyclo[3.1.0]Hexan-6-Yl]Ethanoic Acid)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-09-23 Classification: TRANSFERASE Ligands: S6D, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2018-01-17 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 9E1, DMS |
 |
Crystal Structure Of Ask1 Kinase Domain With A Potent Inhibitor (Analog 13)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2018-01-17 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 9E4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2015-03-18 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: MN, 3FE |
 |
Crystal Structure Of Mst3 With A Pyrrolopyrimidine Inhibitor (Pf-06454589).
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2015-03-18 Classification: transferase/transferase inhibitor Ligands: 3JA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2015-03-18 Classification: transferase/transferase inhibitor Ligands: 3JB |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-09-25 Classification: MEMBRANE PROTEIN Ligands: NAG, CL |
 |
Measurement Of Conformational Changes Accompanying Desensitization In An Ionotropic Glutamate Receptor: Structure Of G725C Mutant
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-10-17 Classification: MEMBRANE PROTEIN Ligands: ZN, GLU |
 |
Measurement Of Conformational Changes Accompanying Desensitization In An Ionotropic Glutamate Receptor: Structure Of S729C Mutant
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-10-17 Classification: MEMBRANE PROTEIN Ligands: GLU |

