Search Count: 23
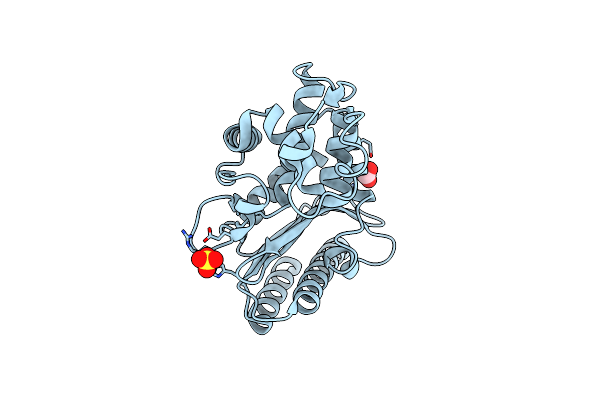 |
High Resolution Structure Of Class A Beta-Lactamase From Bordetella Bronchiseptica Rb50
Organism: Bordetella bronchiseptica rb50
Method: X-RAY DIFFRACTION Release Date: 2024-06-12 Classification: HYDROLASE Ligands: FMT, SO4 |
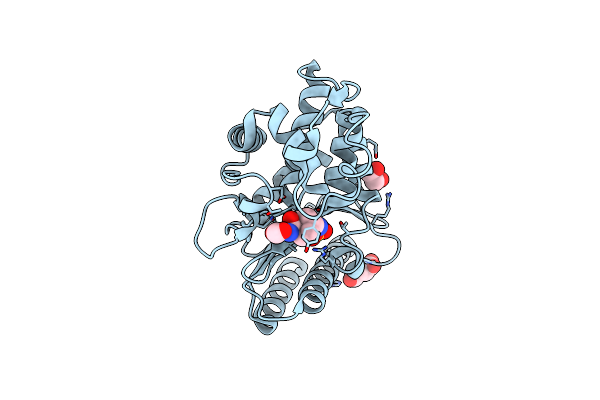 |
Structure Of Class A Beta-Lactamase From Bordetella Bronchiseptica Rb50 In A Complex With Avibactam
Organism: Bordetella bronchiseptica rb50
Method: X-RAY DIFFRACTION Release Date: 2024-06-12 Classification: HYDROLASE Ligands: NXL, SIN, FMT, EDO |
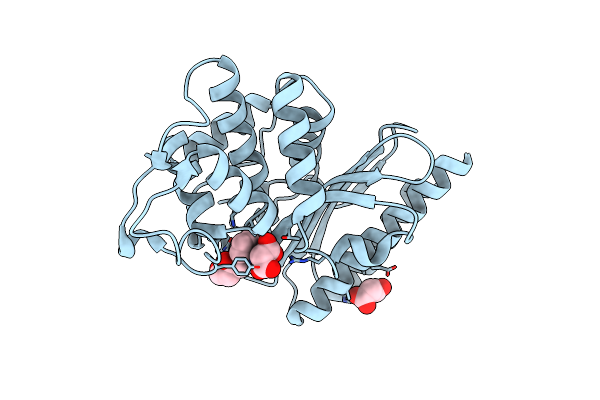 |
Structure Of Class A Beta-Lactamase From Bordetella Bronchiseptica Rb50 In A Complex With Clavulonate
Organism: Bordetella bronchiseptica rb50
Method: X-RAY DIFFRACTION Release Date: 2024-06-12 Classification: HYDROLASE Ligands: MLA, TEM, CL |
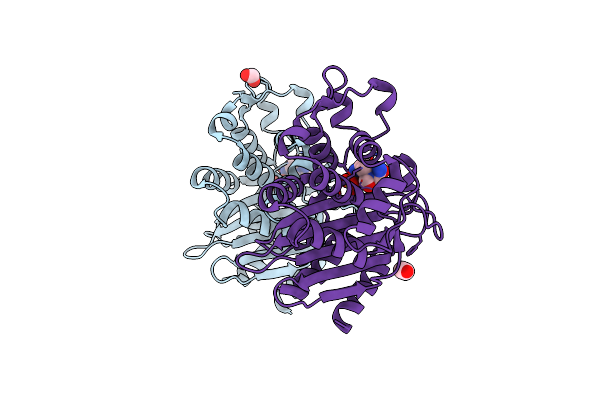 |
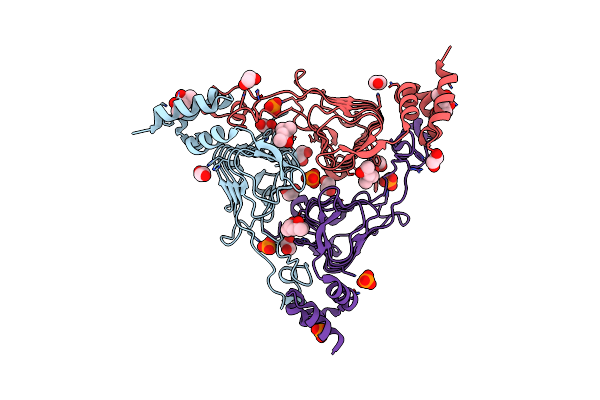
Crystal Structure Of The Beta Lactamase Class A Penp From Bacillus Subtilis In The Complex With The Non-Beta- Lactam Beta-Lactamase Inhibitor Avibactam
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Release Date: 2020-03-25 Classification: HYDROLASE Ligands: NXL, FMT, EDO |
 |
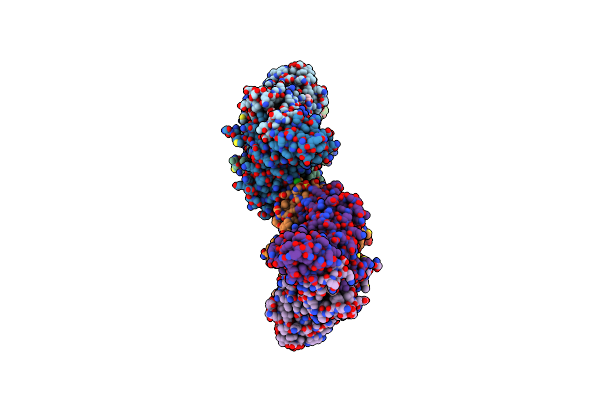
The 2.0 A Crystal Structure Of The Type B Chloramphenicol Acetyltransferase From Vibrio Cholerae
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Release Date: 2019-09-25 Classification: TRANSFERASE Ligands: MPD, EDO, CL, PO4 |
 |
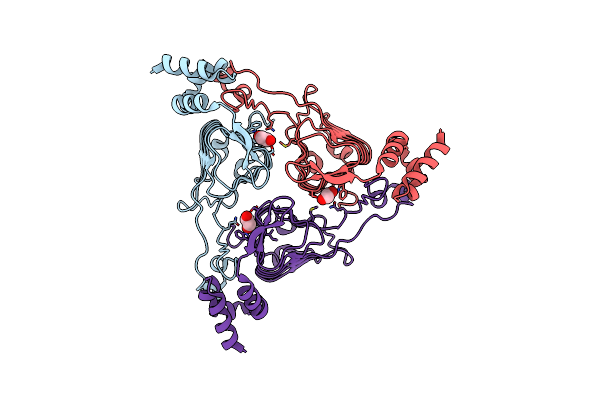
The Crystal Structure Of Chloramphenicol Acetyltransferase-Like Protein From Vibrio Fischeri Es114 In Complex With Taurocholic Acid
Organism: Aliivibrio fischeri (strain atcc 700601 / es114)
Method: X-RAY DIFFRACTION Release Date: 2019-09-25 Classification: TRANSFERASE Ligands: TCH, CL, SO4, ACT, GOL, FMT |
 |
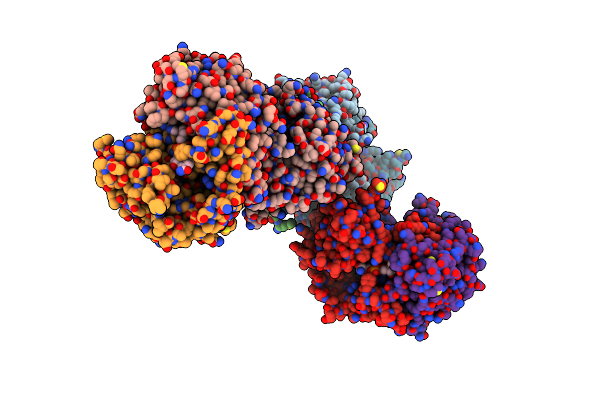
Crystal Structure Of The Type B Chloramphenicol O-Acetyltransferase From Vibrio Vulnificus
Organism: Vibrio vulnificus (strain cmcp6)
Method: X-RAY DIFFRACTION Release Date: 2019-08-14 Classification: TRANSFERASE Ligands: EDO, CL |
 |
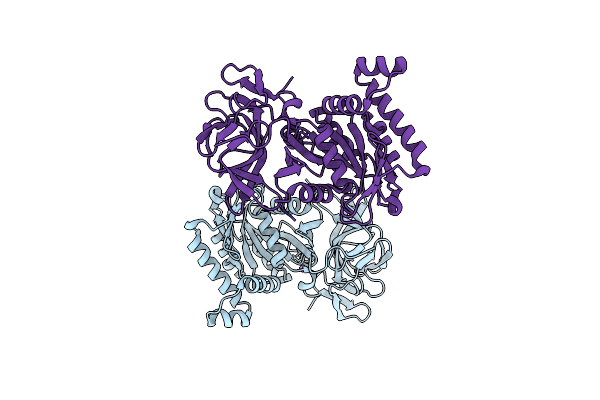
Crystal Structure Of Nad Synthetase (Nade) From Enterococcus Faecalis In Complex With Nad+
Organism: Enterococcus faecalis v583
Method: X-RAY DIFFRACTION Release Date: 2018-02-14 Classification: TRANSFERASE Ligands: NAD |
 |
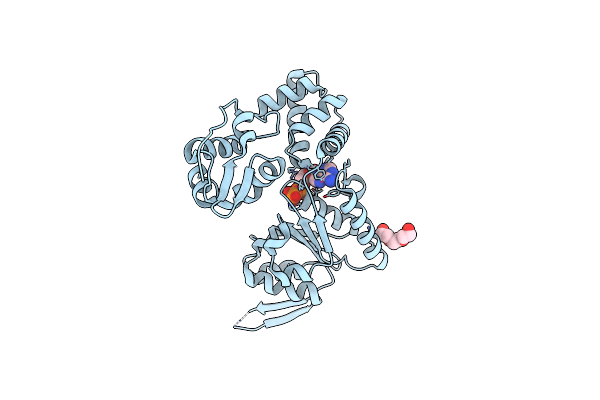
1.83 Angstrom Resolution Crystal Structure Of N-Terminal Fragment (Residues 1-404) Of Elongation Factor G From Enterococcus Faecalis
Organism: Enterococcus faecalis (strain atcc 700802 / v583)
Method: X-RAY DIFFRACTION Release Date: 2017-11-22 Classification: RIBOSOMAL PROTEIN |
 |
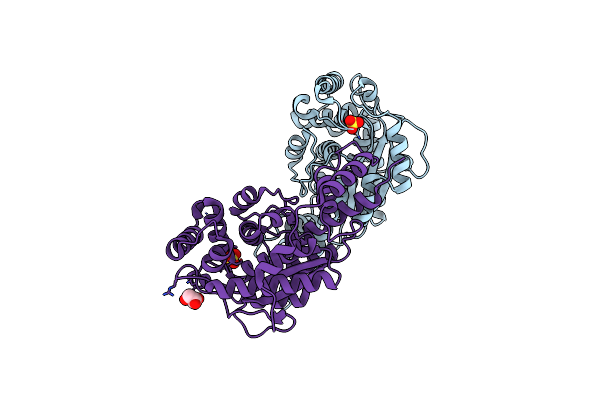
1.88 Angstrom Resolution Crystal Structure Holliday Junction Atp-Dependent Dna Helicase (Ruvb) From Pseudomonas Aeruginosa In Complex With Adp
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Release Date: 2017-11-22 Classification: HYDROLASE Ligands: ADP, PGE |
 |
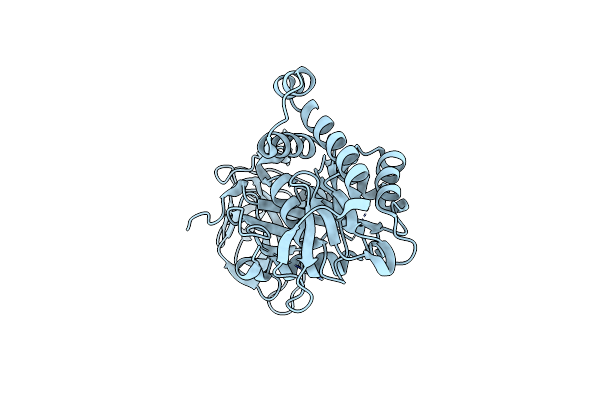
Crystal Structure Of Nicotinate Mononucleotide-5,6-Dimethylbenzimidazole Phosphoribosyltransferase Cobt From Yersinia Enterocolitica
Organism: Yersinia enterocolitica serotype o:8 / biotype 1b (strain nctc 13174 / 8081)
Method: X-RAY DIFFRACTION Release Date: 2017-10-18 Classification: TRANSFERASE Ligands: SO4, CL, GOL |
 |
1.78 Angstrom Resolution Crystal Structure Of N-Terminal Fragment (Residues 1-405) Of Elongation Factor G From Haemophilus Influenzae
Organism: Haemophilus influenzae (strain atcc 51907 / dsm 11121 / kw20 / rd)
Method: X-RAY DIFFRACTION Release Date: 2017-10-18 Classification: RIBOSOMAL PROTEIN Ligands: CL |
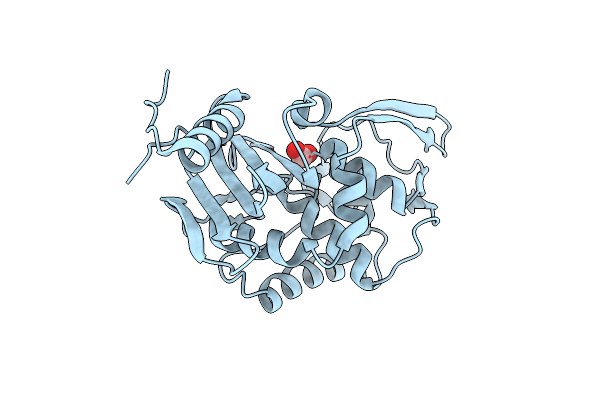 |
1.75 Angstrom Resolution Crystal Structure Of D-Alanyl-D-Alanine Endopeptidase From Enterobacter Cloacae In Complex With Covalently Bound Boronic Acid
Organism: Enterobacter cloacae subsp. cloacae atcc 13047
Method: X-RAY DIFFRACTION Release Date: 2017-10-04 Classification: HYDROLASE Ligands: BO4 |
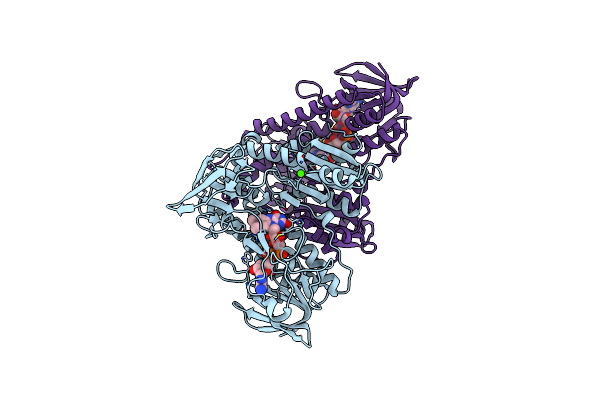 |
1.72 Angstrom Resolution Crystal Structure Of 2-Oxoglutarate Dehydrogenase Complex Subunit Dihydrolipoamide Dehydrogenase From Bordetella Pertussis In Complex With Fad
Organism: Bordetella pertussis (strain tohama i / atcc baa-589 / nctc 13251)
Method: X-RAY DIFFRACTION Release Date: 2017-08-23 Classification: OXIDOREDUCTASE Ligands: FAD, CA |
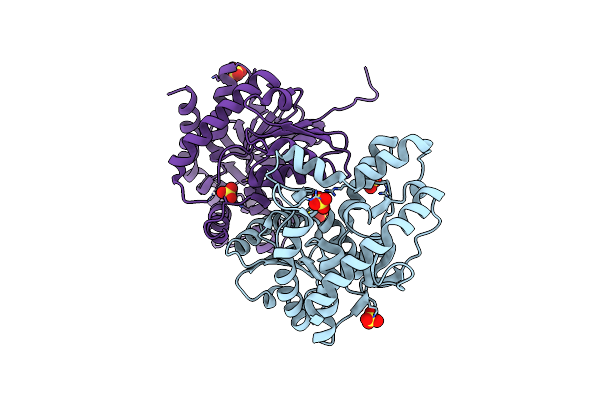 |
2.15 Angstrom Resolution Crystal Structure Of Malate Dehydrogenase From Haemophilus Influenzae
Organism: Haemophilus influenzae (strain atcc 51907 / dsm 11121 / kw20 / rd)
Method: X-RAY DIFFRACTION Release Date: 2017-08-23 Classification: OXIDOREDUCTASE Ligands: SO4 |
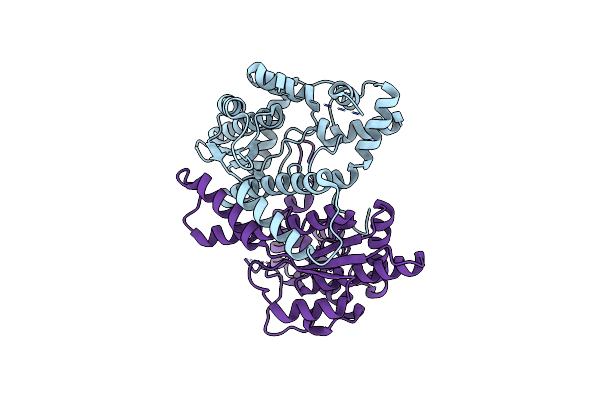 |
Organism: Vibrio fischeri (strain atcc 700601 / es114)
Method: X-RAY DIFFRACTION Release Date: 2017-08-16 Classification: LIGASE |
 |
The Crystal Structure Of Thioredoxin Reductase From Streptococcus Pyogenes Mgas5005
Organism: Streptococcus pyogenes
Method: X-RAY DIFFRACTION Release Date: 2017-03-15 Classification: OXIDOREDUCTASE Ligands: FAD, PO4 |
 |
The Crystal Structure Of Chloramphenicol Acetyltransferase From Vibrio Fischeri Es114
Organism: Vibrio fischeri (strain atcc 700601 / es114)
Method: X-RAY DIFFRACTION Release Date: 2017-03-08 Classification: TRANSFERASE Ligands: MES, MG, ACT, CL, FMT |
 |
The Crystal Structure Of Nitroreductase A From Vibrio Parahaemolyticus Rimd 2210633
Organism: Vibrio parahaemolyticus serotype o3:k6 (strain rimd 2210633)
Method: X-RAY DIFFRACTION Release Date: 2017-03-01 Classification: OXIDOREDUCTASE Ligands: FMN, CL, GOL |
 |
Crystal Structure Of 5,10-Methylenetetrahydrofolate Reductase Metf From Haemophilus Influenzae
Organism: Haemophilus influenzae (strain atcc 51907 / dsm 11121 / kw20 / rd)
Method: X-RAY DIFFRACTION Release Date: 2017-02-22 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, ACY, EDO, FMT |

