Search Count: 28
 |
Set Domain Of Histone-Lysine N-Methyltransferase Nsd2 In Complex With Selective Inhibitor
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-04-30 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: ZN, A1A0M |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-12-14 Classification: PROTEIN BINDING Ligands: DMS |
 |
First-In-Class Small Molecule Inhibitors Of Polycomb Repressive Complex 1 (Prc1) Ring Domain
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2021-06-16 Classification: GENE REGULATION Ligands: ZN, U9E |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-04-14 Classification: LIGASE Ligands: ZN, EPE, GOL |
 |
Inhibitor Compound-Induced Confrontational Change In Ring1B-Bmi1 Domain Structure
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2021-04-14 Classification: LIGASE Ligands: ZN |
 |
Nmr Solution Structure Of Monomeric Ccl5 In Complex With A Doubly-Sulfated N-Terminal Segment Of Ccr5
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2018-04-18 Classification: PROTEIN BINDING |
 |
Nmr Solution Structure Of Bacteriocin Bacsp222 From Staphylococcus Pseudintermedius 222
Organism: Staphylococcus pseudintermedius
Method: SOLUTION NMR Release Date: 2017-10-25 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2016-11-16 Classification: TRANSCRIPTION |
 |
Solution Structure Of The Ims Domain Of The Mitochondrial Import Protein Tim21 From S. Cerevisiae
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2014-10-29 Classification: PROTEIN TRANSPORT |
 |
Solution Structure Of The Mitochondrial Translocator Protein (Tspo) In Complex With Its High-Affinity Ligand Pk11195
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 2014-04-02 Classification: MEMBRANE PROTEIN Ligands: PKA |
 |
High Resolution Structure And Dynamics Of Cspina Parvulin At Physiological Temperature
Organism: Cenarchaeum symbiosum
Method: SOLUTION NMR Release Date: 2013-12-04 Classification: ISOMERASE |
 |
Refined Solution Structure And Dynamics Of First Catalytic Cysteine Half-Domain From Mouse E1 Enzyme
|
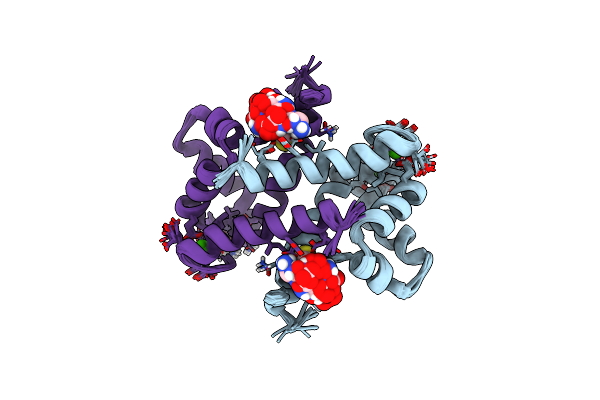 |
Solution Structure And Dynamics Of Human S100A1 Protein Modified At Cysteine 85 With Homocysteine Disulfide Bond Formation In Calcium Saturated Form
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2013-02-20 Classification: METAL BINDING PROTEIN Ligands: HCS, CA |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2013-02-20 Classification: METAL BINDING PROTEIN Ligands: CA |
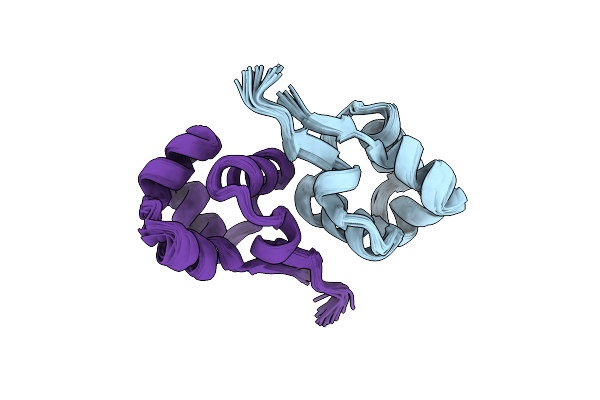 |
Organism: Enterococcus faecalis
Method: SOLUTION NMR Release Date: 2013-02-20 Classification: DNA BINDING PROTEIN |
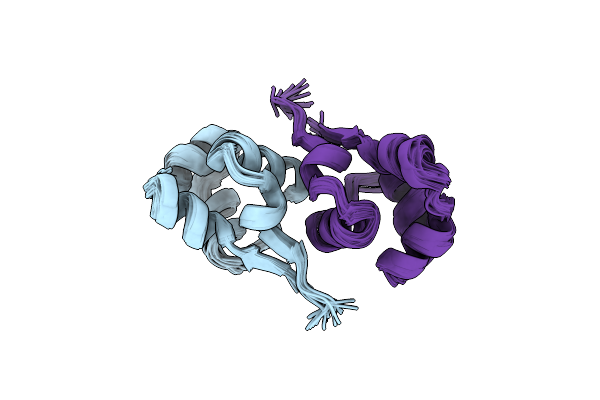 |
Organism: Enterococcus faecalis
Method: SOLUTION NMR Release Date: 2013-02-20 Classification: DNA BINDING PROTEIN |
 |
Noe-Based 3D Structure Of The Predissociated Homodimer Of Cylr2 In Equilibrium With Monomer At 266K (-7 Celsius Degrees)
Organism: Enterococcus faecalis
Method: SOLUTION NMR Release Date: 2013-02-20 Classification: DNA BINDING PROTEIN |
 |
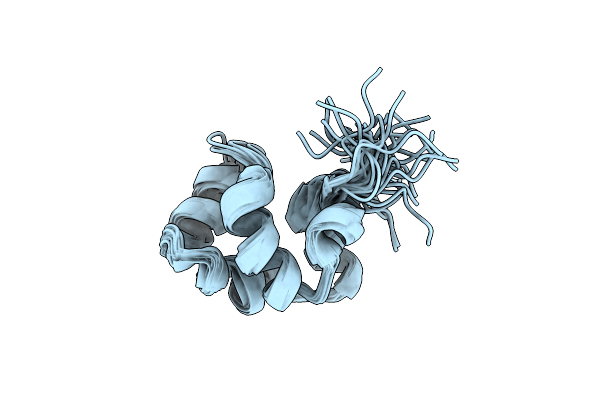
Noe-Based 3D Structure Of The Monomer Of Cylr2 In Equilibrium With Predissociated Homodimer At 266K (-7 Celsius Degrees)
Organism: Enterococcus faecalis
Method: SOLUTION NMR Release Date: 2013-02-20 Classification: DNA BINDING PROTEIN |
 |
Noe-Based 3D Structure Of The Monomeric Intermediate Of Cylr2 At 262K (-11 Celsius Degrees)
Organism: Enterococcus faecalis
Method: SOLUTION NMR Release Date: 2013-02-20 Classification: DNA BINDING PROTEIN |
 |
Noe-Based 3D Structure Of The Monomeric Partially-Folded Intermediate Of Cylr2 At 259K (-14 Celsius Degrees)
Organism: Enterococcus faecalis
Method: SOLUTION NMR Release Date: 2013-02-20 Classification: DNA BINDING PROTEIN |

