Search Count: 109
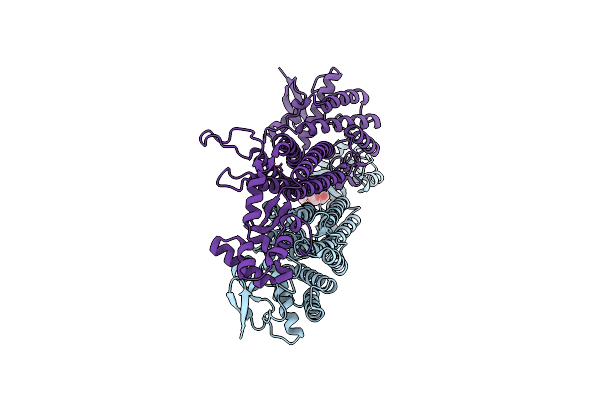 |
Crystal Structure Of Mutant Aspartase From Bacillus Sp. Ym55-1 In The Closed Loop Conformation
Organism: Bacillus sp. ym55-1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-01-15 Classification: LYASE Ligands: PGE, NA |
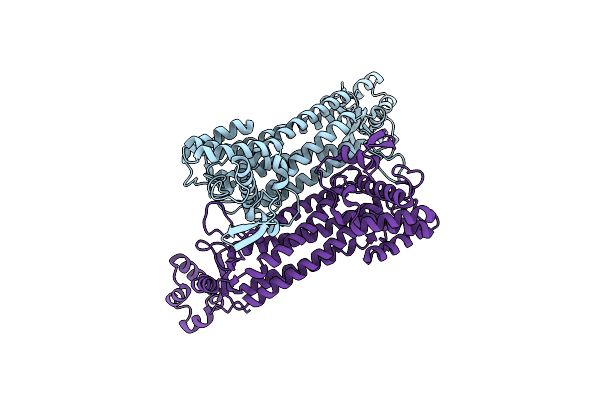 |
Crystal Structure Of Mutant Aspartase From Caenibacillus Caldisaponilyticus In The Closed Loop Conformation
Organism: Caenibacillus caldisaponilyticus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-01-15 Classification: LYASE |
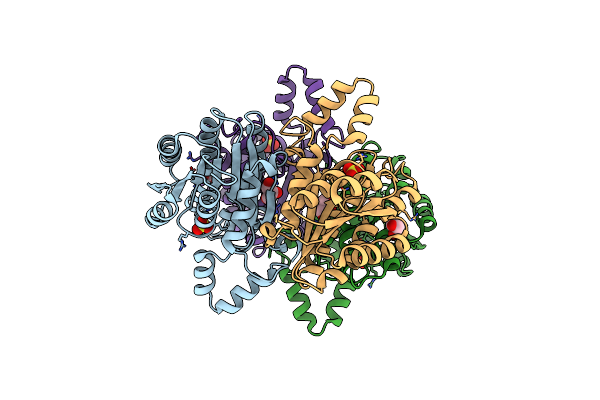 |
Crystal Structure Of Alcohol Dehydrogenase/Ketoreductase Variant From Thermus Thermophilus Apo Form
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: SO4, GOL |
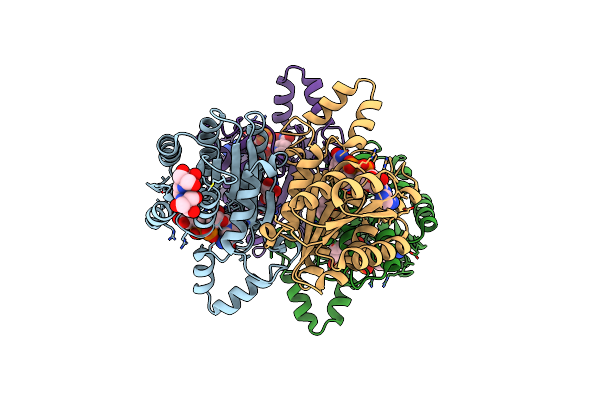 |
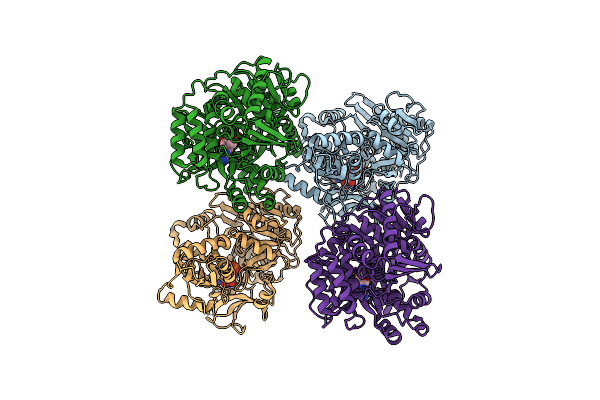
Crystal Structure Of Alcohol Dehydrogenase/Ketoreductase Variant From Thermus Thermophilus Apo Form
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: NAD, B3P, GOL |
 |
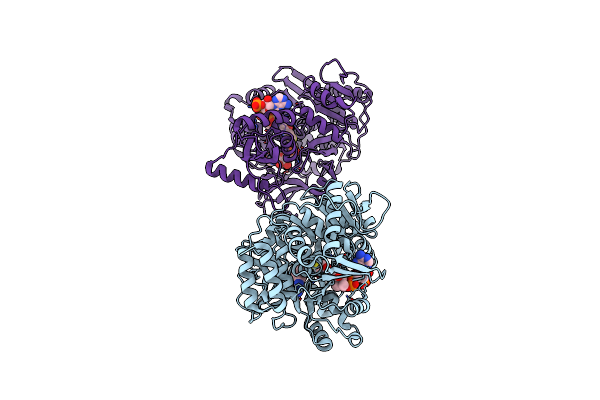
Crystal Structure Of Acyl-Coa Synthetase From Metallosphaera Sedula In Complex With Acetyl-Amp
Organism: Metallosphaera sedula dsm 5348
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-11-15 Classification: LIGASE Ligands: AMP, 6R9 |
 |
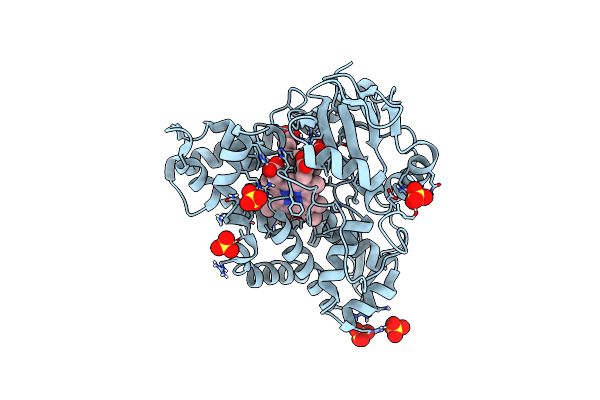
Crystal Structure Of Acyl-Coa Synthetase From Metallosphaera Sedula In Complex With Coenzyme A And Acetyl-Amp
Organism: Metallosphaera sedula dsm 5348
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2023-11-15 Classification: LIGASE Ligands: 6R9, COA |
 |
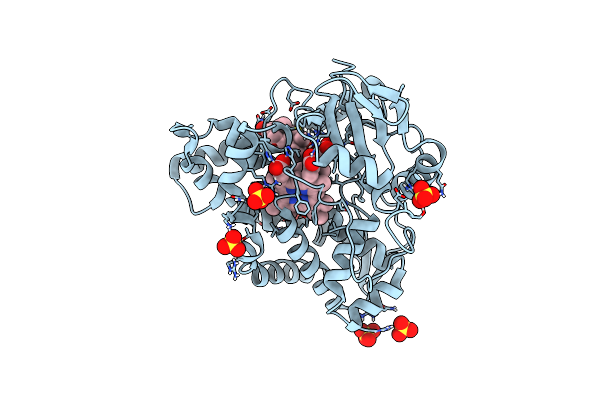
Crystal Structure Of Cyp109A2 From Bacillus Megaterium Bound With Putative Ligands Hexanoic Acid And Octanoic Acid
Organism: Priestia megaterium dsm 319
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: HEB, 6NA, OCA, SO4 |
 |
Crystal Structure Of Cyp109A2 From Bacillus Megaterium Bound With Testosterone And Putative Ligand 4,6-Dimethyloctanoic Acid
Organism: Priestia megaterium dsm 319
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: HEB, W2L, TES, SO4 |
 |
Thermostable Omega Transaminase Pjta-R6 Variant W58G Engineered For Asymmetric Synthesis Of Enantiopure Bulky Amines
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-09-01 Classification: TRANSFERASE Ligands: PLP, SIN |
 |
Thermostable Omega Transaminase Pjta-R6 Variant W58M/F86L/R417L Engineered For Asymmetric Synthesis Of Enantiopure Bulky Amines
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-09-01 Classification: TRANSFERASE Ligands: PMP, SIN |
 |
Crystal Structure Of Atp-Dependent Caprolactamase From Pseudomonas Jessenii
Organism: Pseudomonas jessenii
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2021-04-21 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of Human Steroid Carrier Protein Sl (Scp-2L) Mutant A100C
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2021-03-03 Classification: OXIDOREDUCTASE Ligands: OXN, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2021-03-03 Classification: OXIDOREDUCTASE Ligands: OXN, SO4 |
 |
Organism: Bacillus amyloliquefaciens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2021-02-17 Classification: LIGASE Ligands: GOL, SO4 |
 |
Organism: Bacillus amyloliquefaciens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2021-02-17 Classification: LIGASE Ligands: GOL, SO4 |
 |
Organism: Bacillus amyloliquefaciens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-02-17 Classification: LIGASE |
 |
Organism: Bacillus amyloliquefaciens, Hirudo medicinalis
Method: X-RAY DIFFRACTION Release Date: 2021-02-17 Classification: LIGASE Ligands: GOL, TAR |
 |
Organism: Bacillus amyloliquefaciens, Hirudo medicinalis
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2021-02-17 Classification: LIGASE Ligands: GOL, SO4, PGE |
 |
Organism: Bacillus amyloliquefaciens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2021-02-17 Classification: LIGASE Ligands: HIS, AKR, NA, CL |
 |
Crystal Structure Of Cyp154C5 From Nocardia Farcinica In Complex With 5Alpha-Androstan-3-One
Organism: Nocardia farcinica ifm 10152
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-11-18 Classification: OXIDOREDUCTASE Ligands: HEM, NQ8, CL |

