Search Count: 14
 |
Organism: Muromegalovirus
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2023-10-11 Classification: STRUCTURAL PROTEIN Ligands: BU3, MLI |
 |
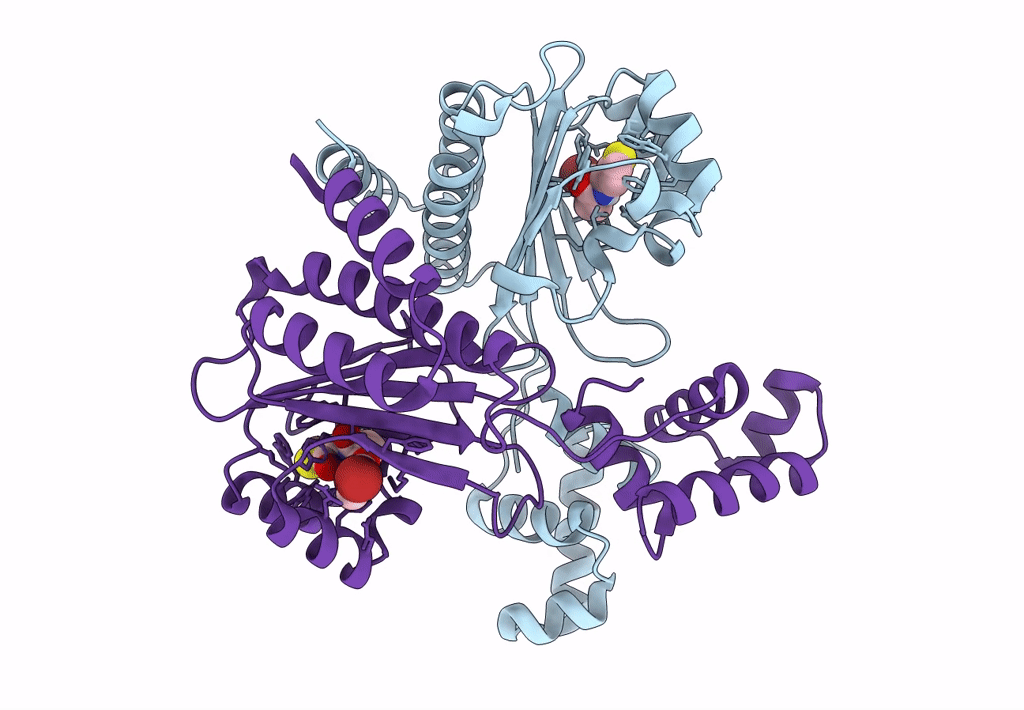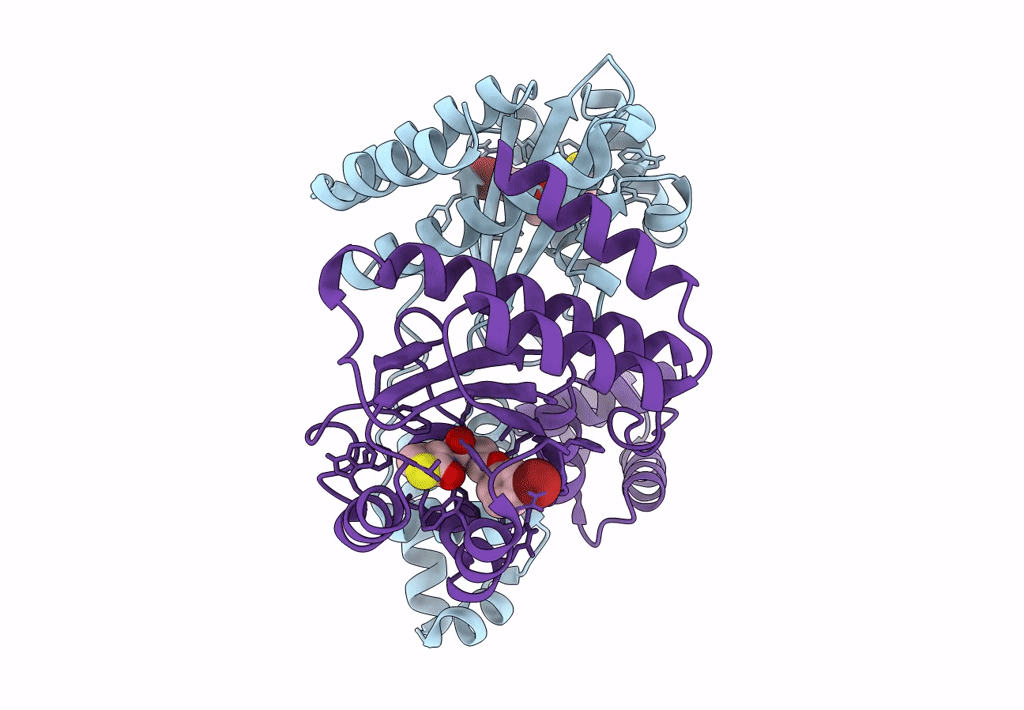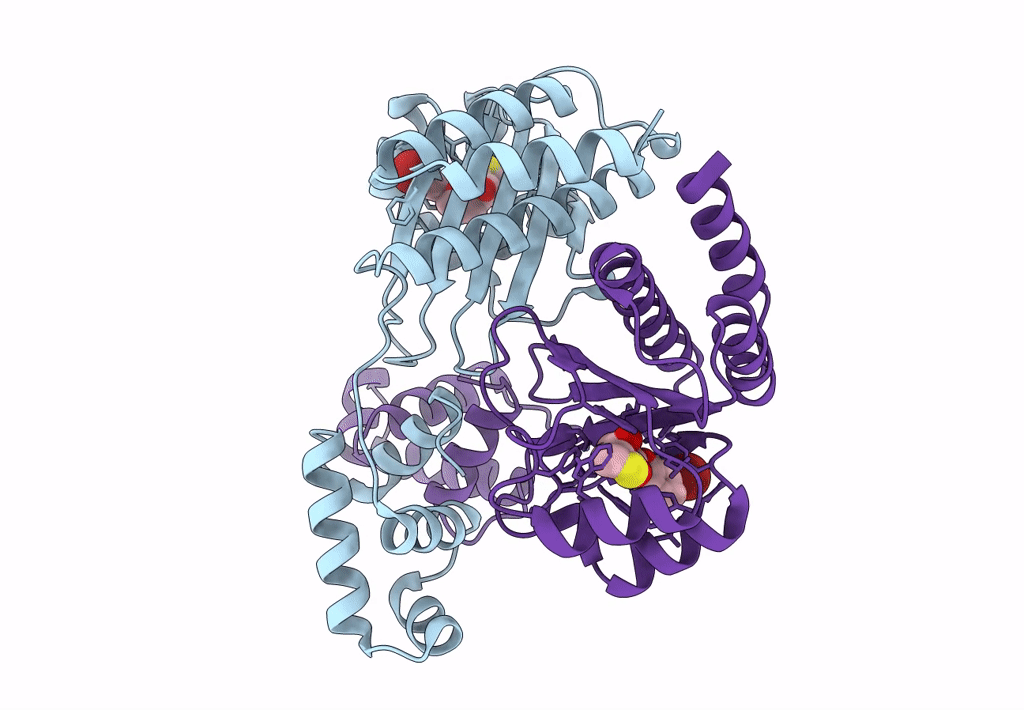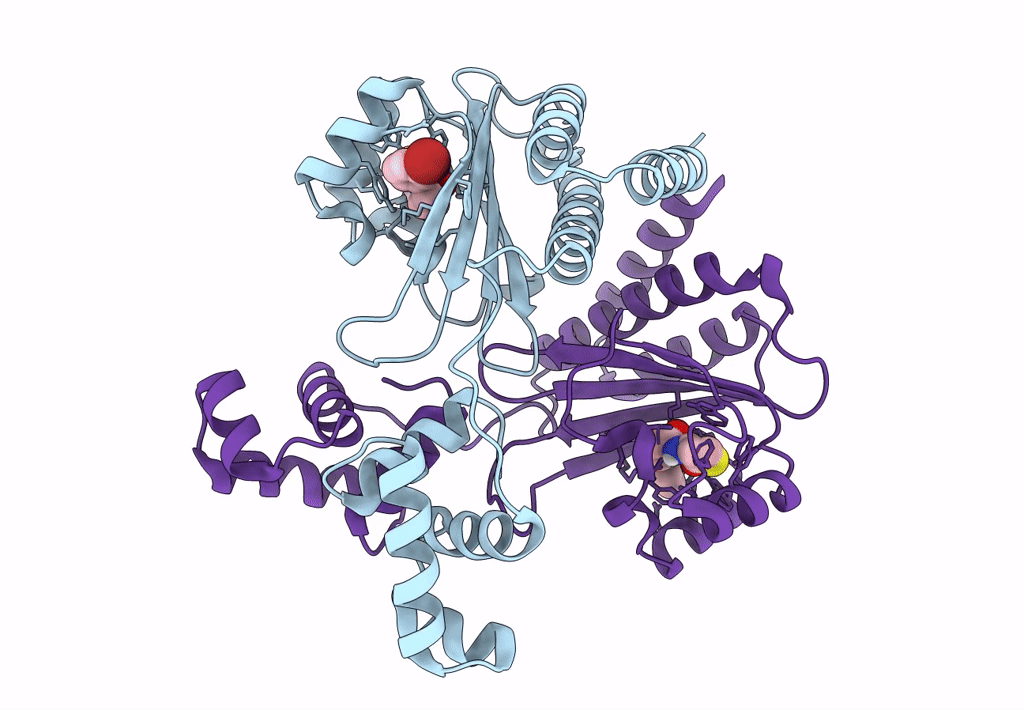
Fusion Construct Of Pqse And Rhlr In Complex With The Synthetic Antagonist Mbtl
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:3.46 Å Release Date: 2022-12-14 Classification: TRANSCRIPTION Ligands: FE, K5G |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-12-14 Classification: HYDROLASE Ligands: FE, CAC, BEZ |
 |
Pross Optimitzed Variant Of Rhlr (75 Mutations) In Complex With The Synthetic Antagonist Mbtl
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-12-14 Classification: TRANSCRIPTION Ligands: K5G |
 |
Pross Optimitzed Variant Of Rhlr (75 Mutations) In Complex With Native Autoinducer C4-Hsl
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2022-12-14 Classification: TRANSCRIPTION Ligands: HL4 |
 |
Pross Optimitzed Variant Of Rhlr (61 Mutations) In Complex With The Synthetic Antagonist Mbtl
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2022-12-14 Classification: TRANSCRIPTION Ligands: K5G |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2022-12-14 Classification: TRANSCRIPTION Ligands: FE, K5G |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2022-12-14 Classification: SIGNALING PROTEIN Ligands: FE, HL4 |
 |
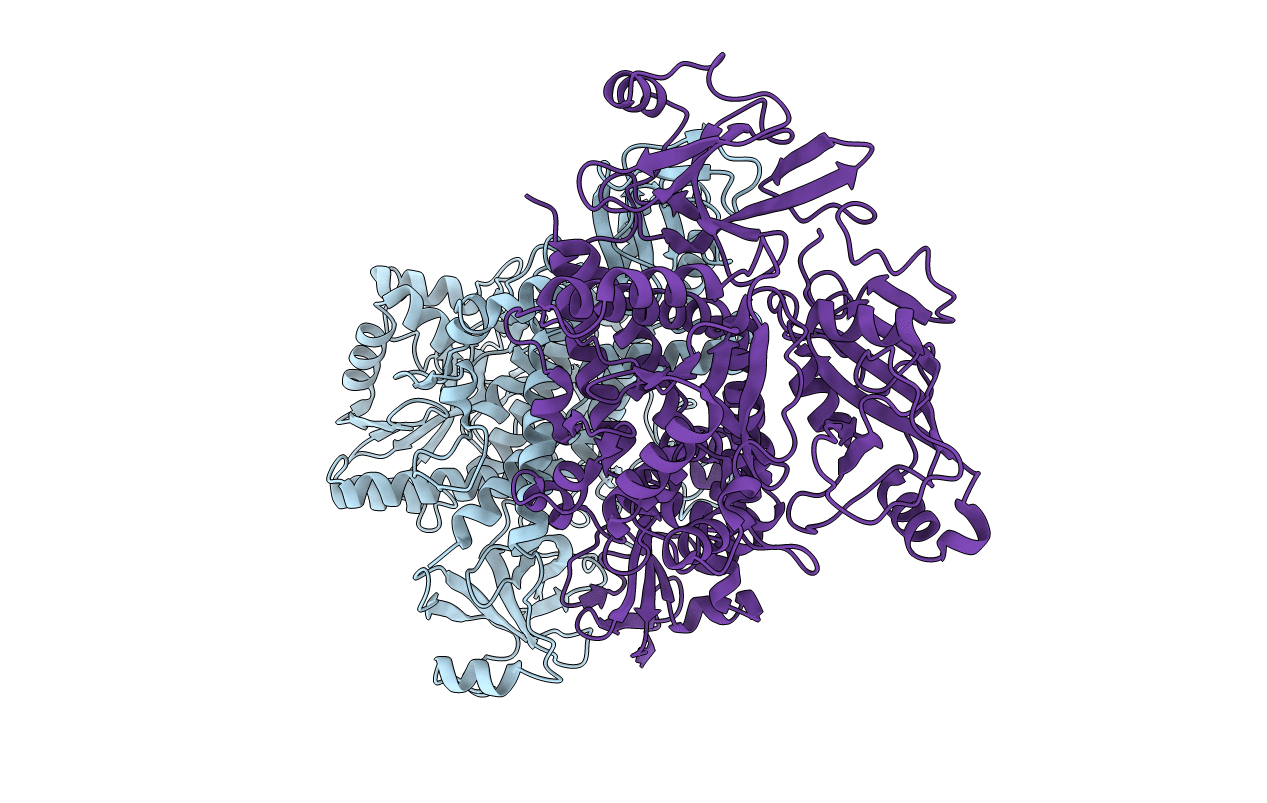
Crystal Structure Of Full-Length Cnfy (C866S) From Yersinia Pseudotuberculosis
Organism: Yersinia pseudotuberculosis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-12-30 Classification: TOXIN Ligands: SO4, CL |
 |
Crystal Structure Of Cnfy From Yersinia Pseudotuberculosis - N-Terminal Fragment Comprising Residues 1-704
Organism: Yersinia pseudotuberculosis
Method: X-RAY DIFFRACTION Resolution:3.28 Å Release Date: 2020-12-30 Classification: TOXIN |
 |
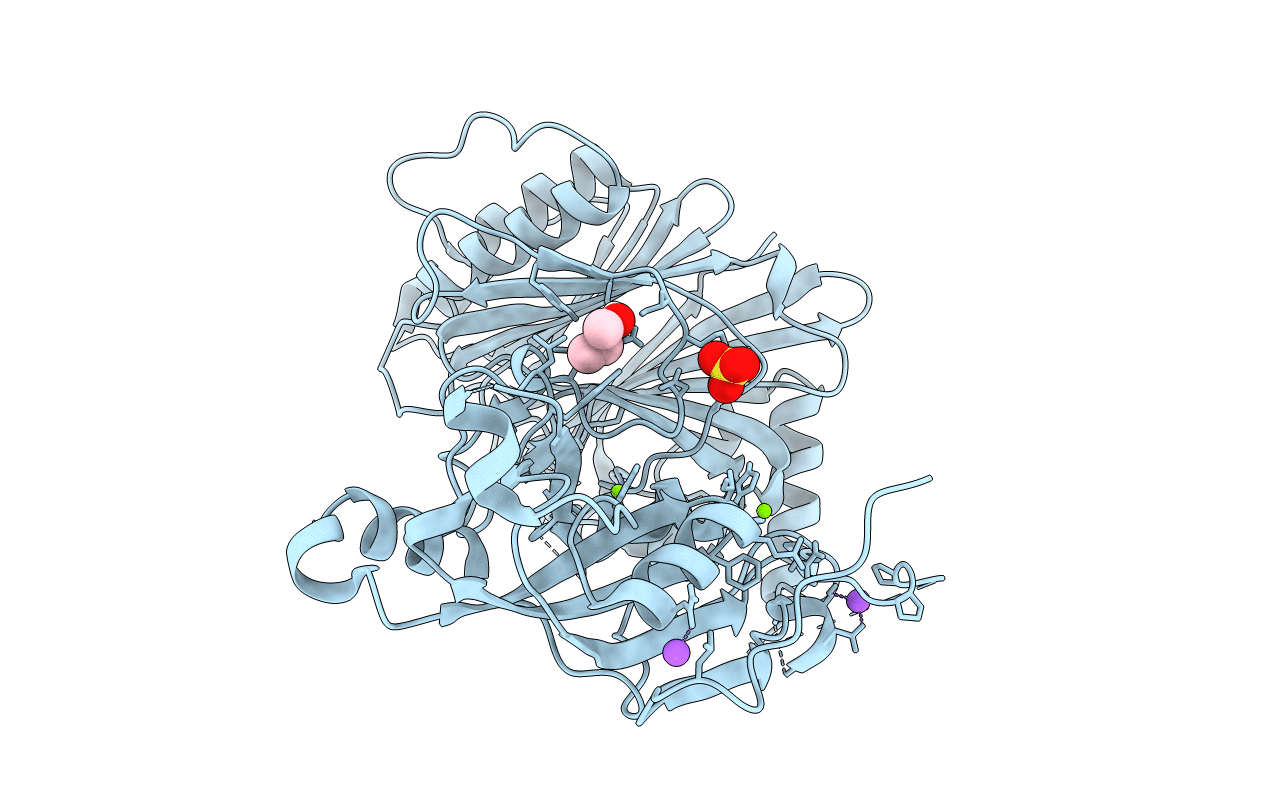
Crystal Structure Of The C-Terminal Domain Of Cnfy From Yersinia Pseudotuberculosis
Organism: Yersinia pseudotuberculosis
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2020-12-30 Classification: TOXIN Ligands: MG |
 |
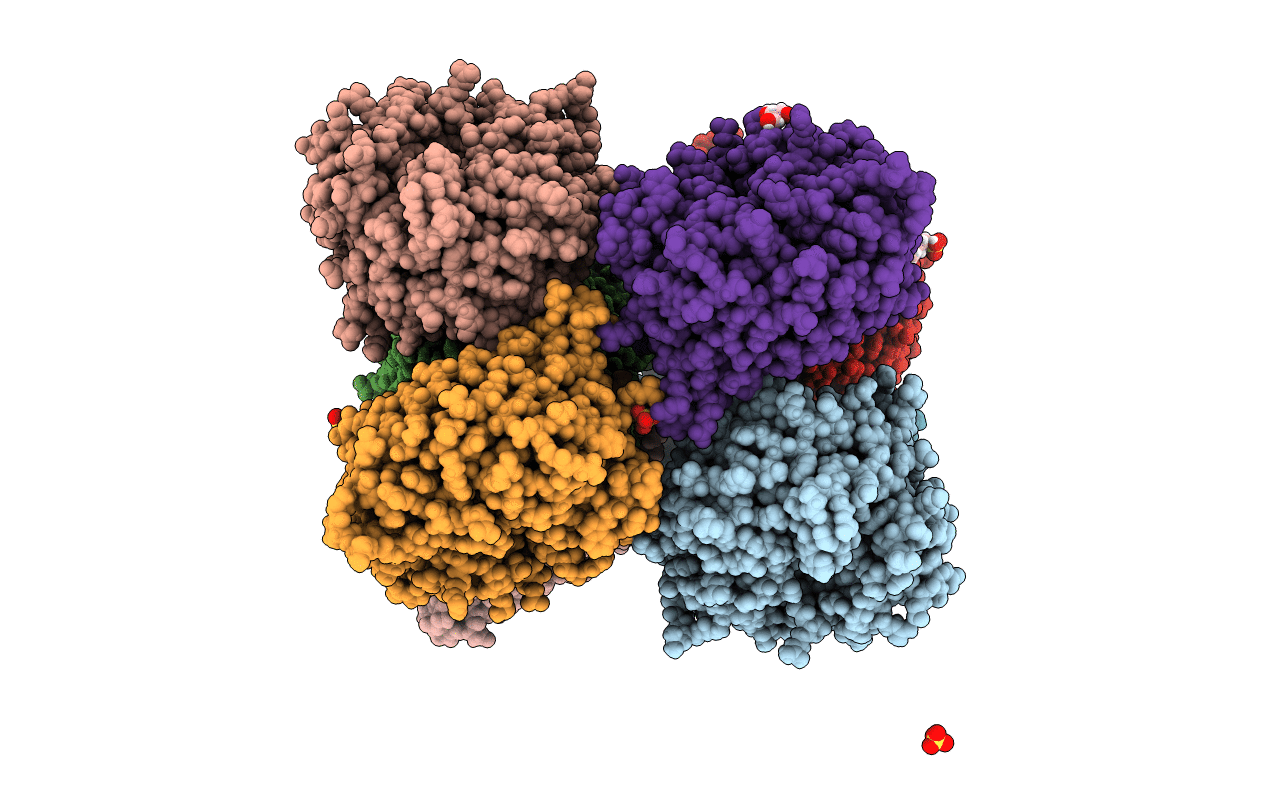
Organism: Yersinia pseudotuberculosis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-12-30 Classification: TOXIN Ligands: BU3, MG, NA, CL, SO4 |
 |
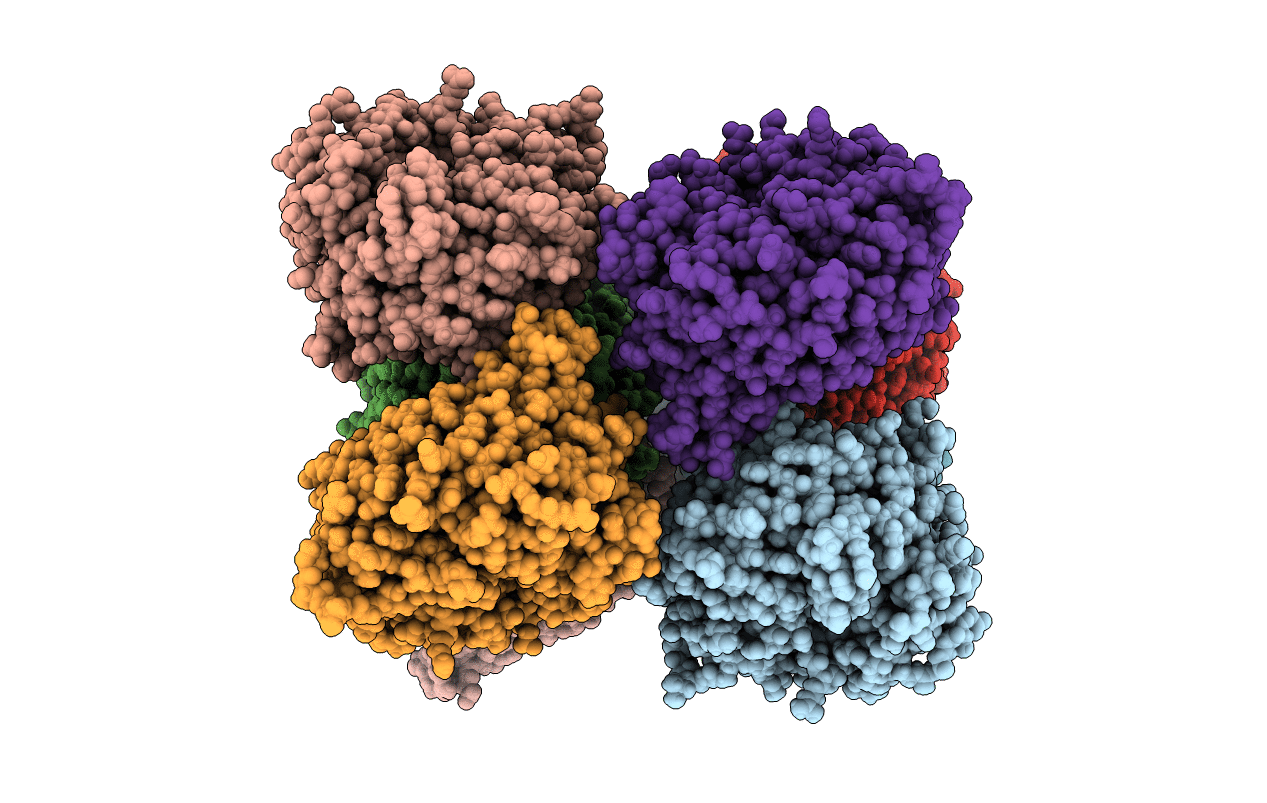
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2020-04-22 Classification: UNKNOWN FUNCTION Ligands: SO4, GOL, MPO |
 |
Heme D1 Biosynthesis Associated Protein Nirf In Complex With Dihydro-Heme D1
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2020-04-22 Classification: UNKNOWN FUNCTION Ligands: DHE |

