Search Count: 42
 |
Organism: Geobacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2024-10-16 Classification: LYASE |
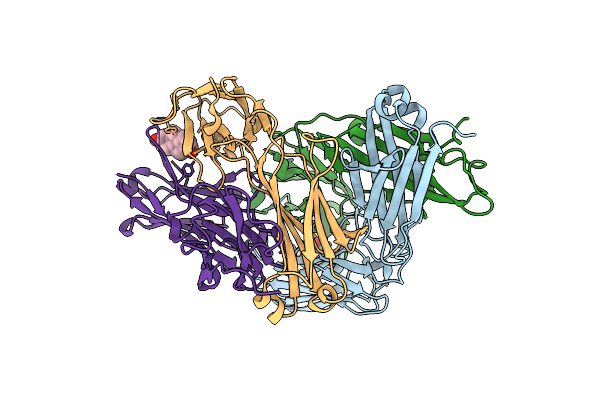 |
Crystal Structure Of The Anti-Testosterone Fab In Complex With Testosterone
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-09-18 Classification: IMMUNE SYSTEM Ligands: TES |
 |
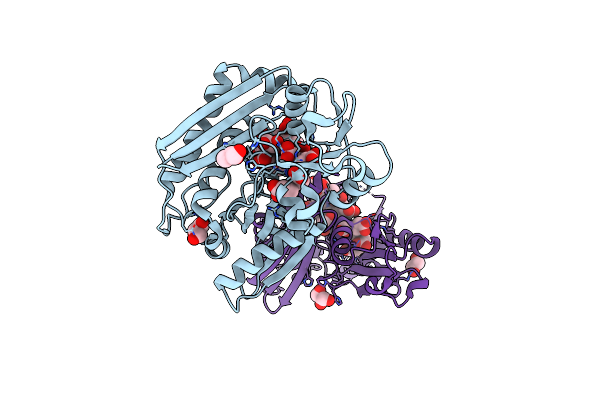
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-09-18 Classification: IMMUNE SYSTEM |
 |
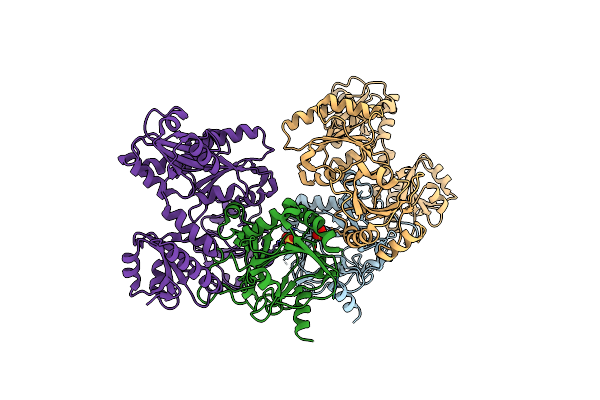
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-11-01 Classification: TRANSFERASE Ligands: ZWW, GOL, EDO |
 |
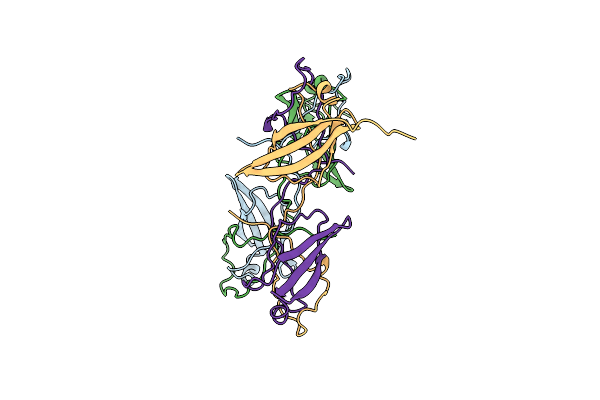
Crystal Structure Of The Heterodimeric Human C-P4H-Ii With Truncated Alpha Subunit (C-P4H-Ii Delta281)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.85 Å Release Date: 2022-11-09 Classification: HYDROLASE Ligands: SO4 |
 |
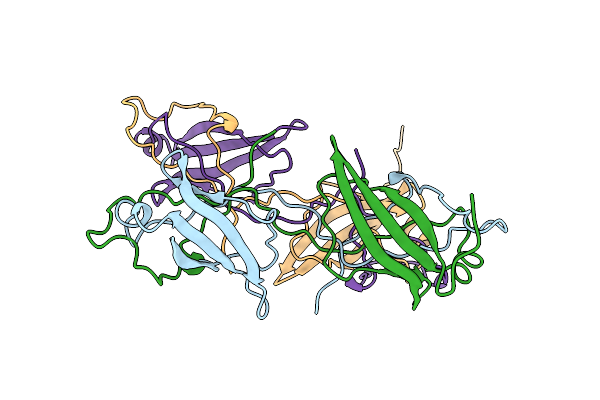
Crystal Structure Of Se-Met Labelled Mce Domain Of Mce4A From Mycobacterium Tuberculosis H37Rv
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:3.61 Å Release Date: 2021-08-25 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of Mce Domain Of Mce4A From Mycobacterium Tuberculosis H37Rv
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2021-08-25 Classification: TRANSPORT PROTEIN |
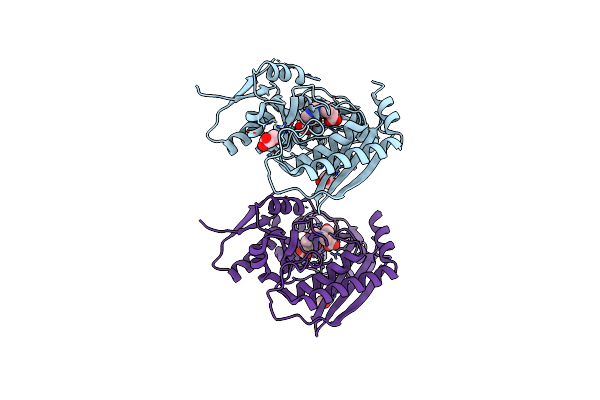 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-02-17 Classification: TRANSFERASE Ligands: DPM, GOL |
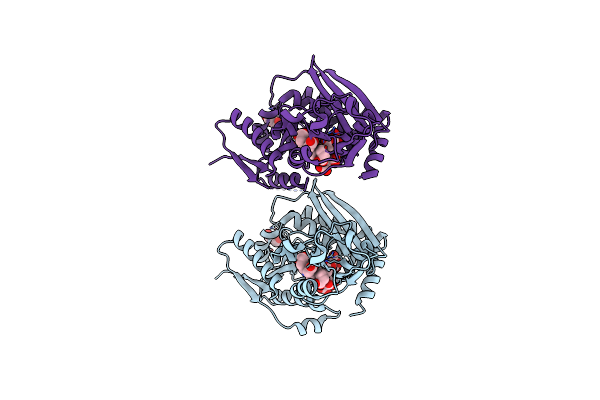 |
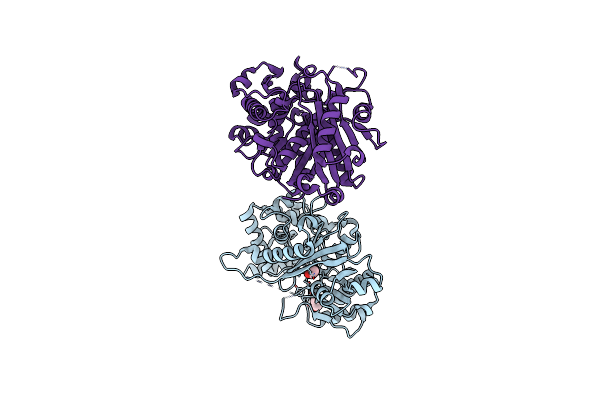
Human Porphobilinogen Deaminase R173W Mutant Crystallized In The Es2 Intermediate State
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-02-17 Classification: TRANSFERASE Ligands: 7J8, GOL |
 |
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-01-02 Classification: TRANSFERASE Ligands: GOL, ACT |
 |
Crystal Structure Of The Zebrafish Peroxisomal Scp2-Thiolase (Type-1) In Complex With Coa
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2019-01-02 Classification: TRANSFERASE Ligands: ACT, GOL, COA |
 |
Crystal Structure Of The Zebrafish Peroxisomal Scp2-Thiolase (Type-1) In Complex With Coa And Octanoyl-Coa
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2019-01-02 Classification: TRANSFERASE Ligands: GOL, CO8, COA |
 |
Crystal Structure Of Aspergillus Oryzae Catechol Oxidase Complexed With Resorcinol
Organism: Aspergillus oryzae (strain atcc 42149 / rib 40)
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2018-09-19 Classification: OXIDOREDUCTASE Ligands: CU, MAN, NAG, RCO, PEO |
 |
Crystal Structure Of Hexameric Cbs-Cp12 Protein From Bloom-Forming Cyanobacteria, Yb-Derivative At 2.8 A Resolution
Organism: Microcystis aeruginosa pcc 7806
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2018-05-30 Classification: PHOTOSYNTHESIS Ligands: YB, DO3 |
 |
Crystal Structure Of Hexameric Cbs-Cp12 Protein From Bloom-Forming Cyanobacteria At 2.5 A Resolution In P6322 Crystal Form
Organism: Microcystis aeruginosa pcc 7806
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-05-30 Classification: PHOTOSYNTHESIS |
 |
Organism: Microcystis aeruginosa pcc 7806
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-05-16 Classification: PHOTOSYNTHESIS Ligands: CL |
 |
Organism: Rhizobium leguminosarum bv. trifolii
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-06-21 Classification: LYASE |
 |
Organism: Rhizobium leguminosarum bv. trifolii (strain wsm2304), Rhizobium leguminosarum bv. trifolii
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-06-21 Classification: LYASE Ligands: MG, FES |
 |
Organism: Rhizobium leguminosarum bv. trifolii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-06-21 Classification: LYASE Ligands: FES, MG |
 |
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-03-23 Classification: LYASE Ligands: GOL, FMT |

