Search Count: 41
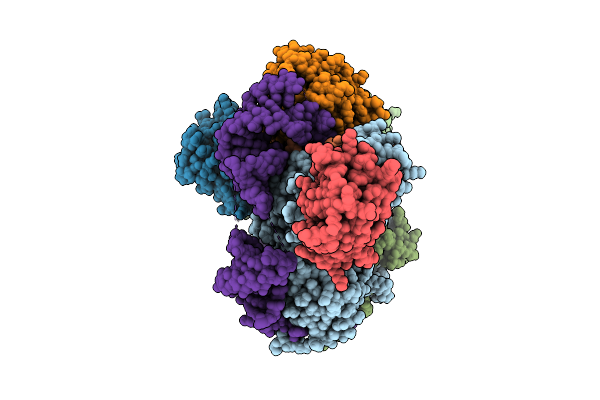 |
Organism: Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: RNA BINDING PROTEIN |
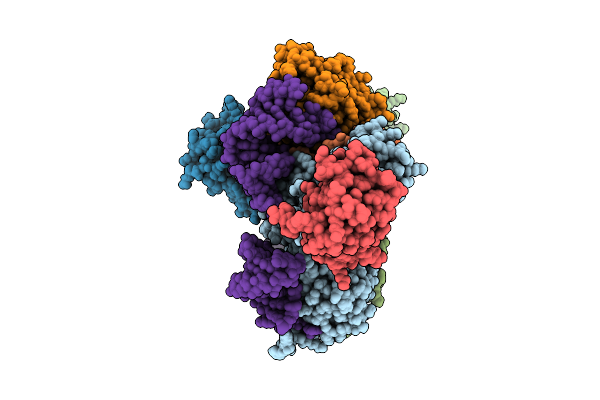 |
Organism: Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: RNA BINDING PROTEIN |
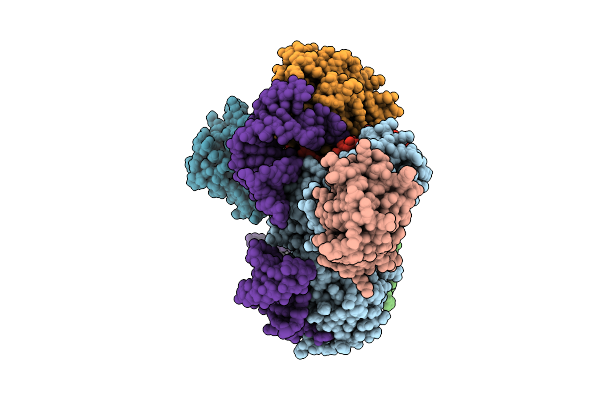 |
Organism: Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: RNA BINDING PROTEIN |
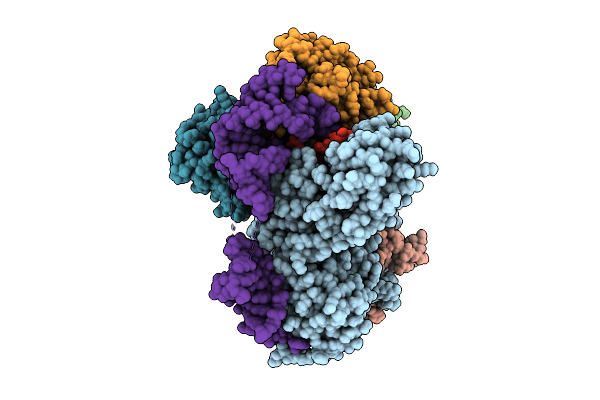 |
Organism: Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: RNA BINDING PROTEIN |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Noncovalently Bound Inhibitor C5N17A
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: VIRAL PROTEIN Ligands: DMS, A1IHT, CL, MG |
 |
Crystal Structure Of Sars-Cov-2 Main Protease (Mpro) In Complex With The Noncovalently Bound Inhibitor C5N17B
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: VIRAL PROTEIN Ligands: DMS, A1IHV, IMD |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2025-03-26 Classification: LYASE |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:3.31 Å Release Date: 2024-12-04 Classification: HYDROLASE |
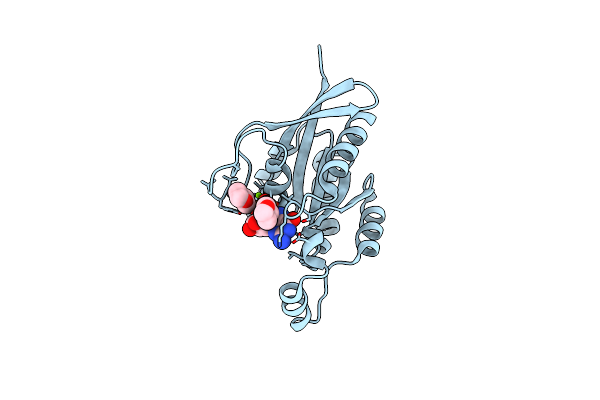 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-06-05 Classification: SIGNALING PROTEIN Ligands: GDP, MG, DIO |
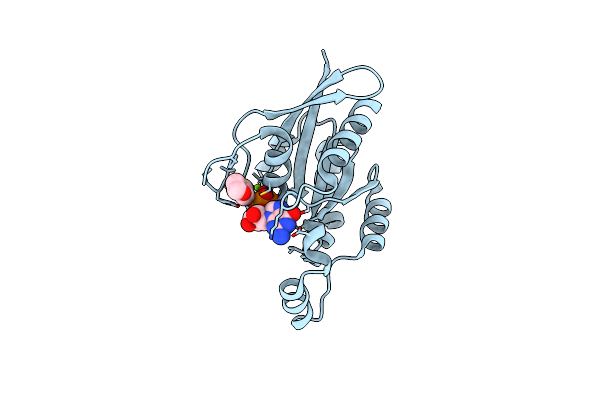 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2024-06-05 Classification: SIGNALING PROTEIN, Hydrolase Ligands: GDP, MG, DIO |
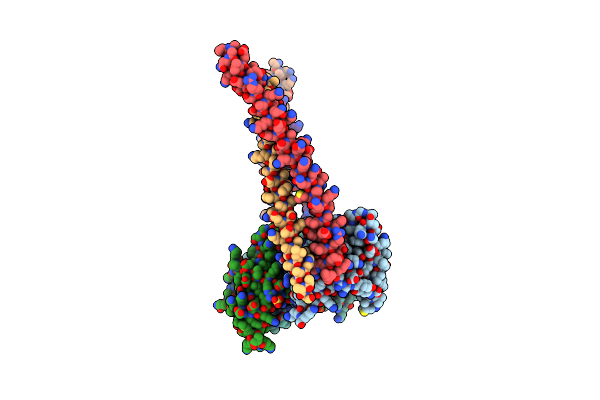 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-05-15 Classification: APOPTOSIS Ligands: BME, SO4 |
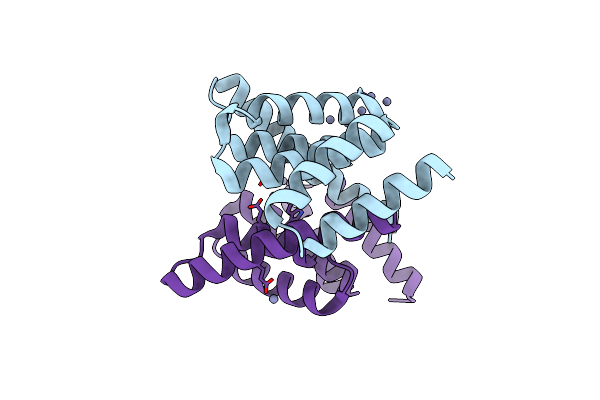 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2023-08-02 Classification: APOPTOSIS Ligands: ZN |
 |
Cryo-Em Structure Of Sars-Cov-2 Receptor Binding Domain In Complex With K202.B Bispecific Antibody
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-07-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Notothenia coriiceps
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2023-07-12 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2023-07-05 Classification: APOPTOSIS Ligands: BME, PEG, NA |
 |
Cryo-Em Structure Of Sars-Cov-2 Spike In Complex With K202.B Bispecific Antibody
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2022-12-07 Classification: HYDROLASE Ligands: R28 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-12-07 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: R28, EDO, CIT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2022-11-30 Classification: TRANSFERASE Ligands: E0X, CL, DMS |
 |
Organism: Mus musculus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2022-06-01 Classification: IMMUNE SYSTEM |

