Search Count: 18
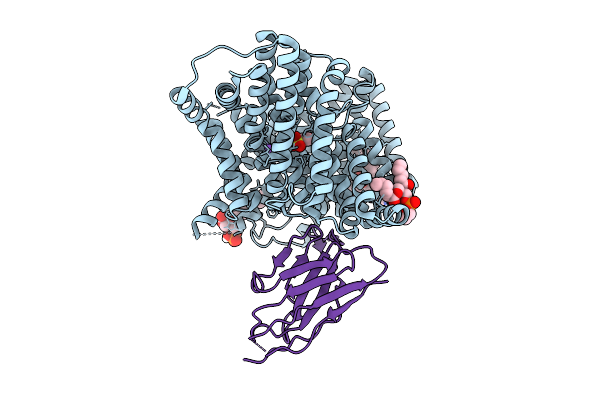 |
Cryo-Em Structure Of The Isethionate Trap Transporter Iseqm From Oleidesulfovibrio Alaskensis With Bound Isethionate
Organism: Oleidesulfovibrio alaskensis g20, Helicobacter pylori
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: PROTEIN BINDING |
 |
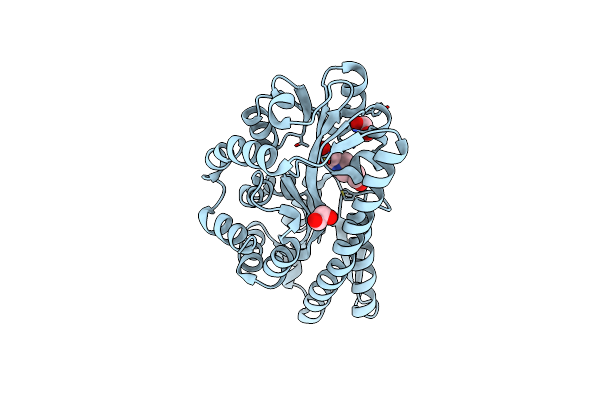
Crystal Structure Of A Mes Bound Substrate Binding Protein (Isep) From An Isethionate Trap Transporter
Organism: Oleidesulfovibrio alaskensis g20
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-08-28 Classification: TRANSPORT PROTEIN Ligands: MES |
 |
Organism: Oleidesulfovibrio alaskensis g20
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-08-28 Classification: TRANSPORT PROTEIN Ligands: EPE, EDO |
 |
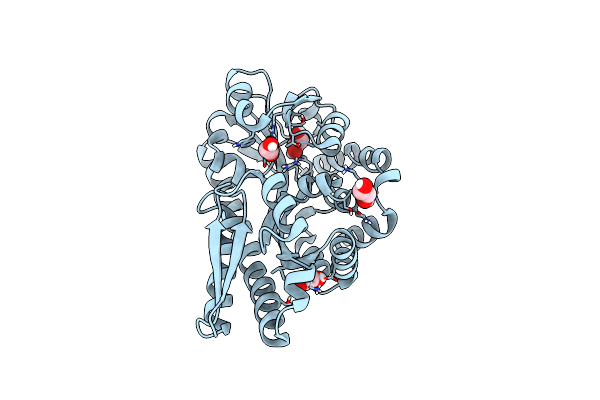
Crystal Structure Of An Isethionate Bound Substrate Binding Protein (Isep) From An Isethionate Trap Transporter
Organism: Oleidesulfovibrio alaskensis g20
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2024-07-10 Classification: TRANSPORT PROTEIN Ligands: 8X3, NO3, EDO |
 |
Apo Crystal Structure Of A Substrate Binding Protein (Isep) From An Isethionate Trap Transporter
Organism: Oleidesulfovibrio alaskensis g20
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2024-06-26 Classification: TRANSPORT PROTEIN Ligands: EDO, CL |
 |
Cryo-Em Structure Of The Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Photobacterium Profundum In A Nanodisc
Organism: Photobacterium profundum ss9, Helicobacter pylori, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-03-15 Classification: TRANSPORT PROTEIN Ligands: OCT, PTY, TWT, D10, NA, TRD, HEX |
 |
Cryo-Em Structure Of The Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Photobacterium Profundum In Amphipol
Organism: Photobacterium profundum ss9, Helicobacter pylori, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-12-21 Classification: TRANSPORT PROTEIN |
 |
Structure Of The Sialic Acid Bound Tripartite Atp-Independent Periplasmic (Trap) Periplasmic Component Siap From Photobacterium Profundum
Organism: Photobacterium profundum
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2022-12-14 Classification: TRANSPORT PROTEIN Ligands: SLB, SO4 |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-09-14 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM |
 |
Fphf, Staphylococcus Aureus Fluorophosphonate-Binding Serine Hydrolases F, Apo Form
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2020-09-16 Classification: HYDROLASE |
 |
Fphf, Staphylococcus Aureus Fluorophosphonate-Binding Serine Hydrolases F, Kt129 Bound
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2020-09-16 Classification: HYDROLASE Ligands: 6WG |
 |
Fphf, Staphylococcus Aureus Fluorophosphonate-Binding Serine Hydrolases F, Kt130 Bound
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2020-09-16 Classification: HYDROLASE Ligands: 6WG |
 |
Fphf, Staphylococcus Aureus Fluorophosphonate-Binding Serine Hydrolases F, Substrate Bound
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2020-09-16 Classification: HYDROLASE Ligands: HE4 |
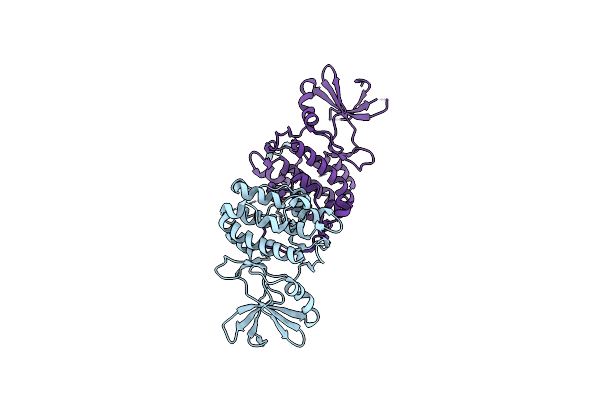 |
Tribbles (Trib1) Pseudokinase Fused To Ccaat-Enhancer Binding Protein (C/Ebpalpha) Degron
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-10-10 Classification: SIGNALING PROTEIN |
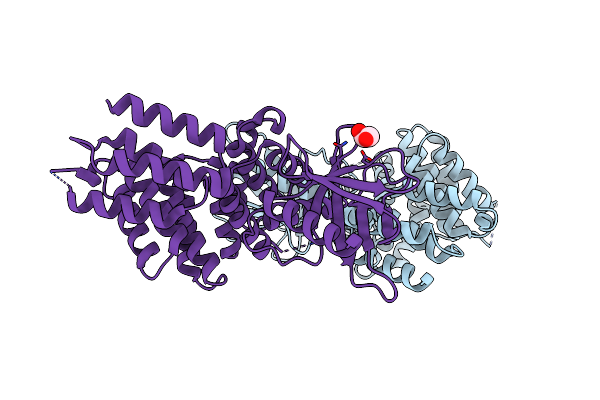 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-03-01 Classification: TRANSFERASE Ligands: GOL |
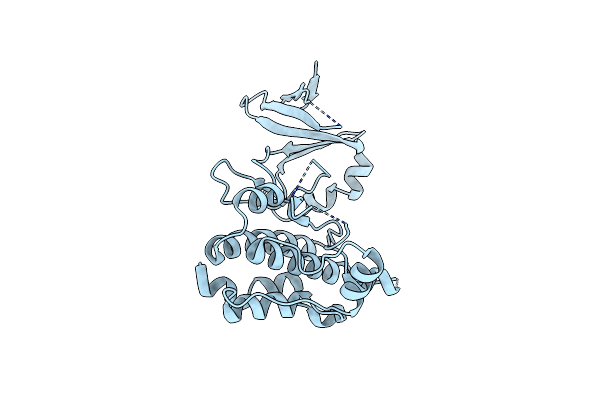 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-11-11 Classification: TRANSFERASE |
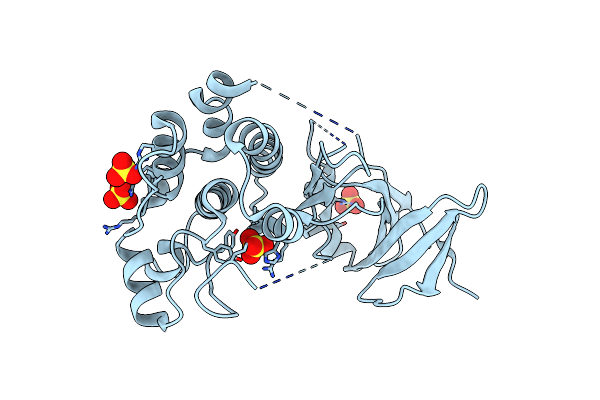 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-11-11 Classification: TRANSFERASE Ligands: SO4 |

