Search Count: 100
 |
Wild Type Apobec3A In Complex With Tt(Fdz)-Hairpin Inhibitor (Crystal Form 1)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2023-09-06 Classification: DNA BINDING PROTEIN Ligands: PO4, ZN |
 |
Wild Type Apobec3A In Complex With Tt(Fdz)-Hairpin Inhibitor (Crystal Form 2)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-09-06 Classification: DNA BINDING PROTEIN Ligands: ZN, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2023-09-06 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN, CL, PO4, IHP |
 |
Zinc-Free Apobec3A (Inactive E72A Mutant) In Complex With Ttc-Hairpin Dna Substrate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2023-09-06 Classification: DNA BINDING PROTEIN Ligands: IHP, CL |
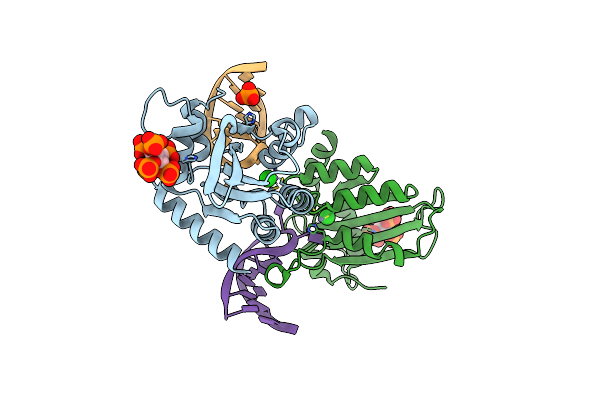 |
Structure Of Apobec3A (E72A Inactive Mutant) In Complex With Ttc-Hairpin Dna Substrate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2023-09-06 Classification: DNA BINDING PROTEIN Ligands: IHP, ZN, CL, PO4 |
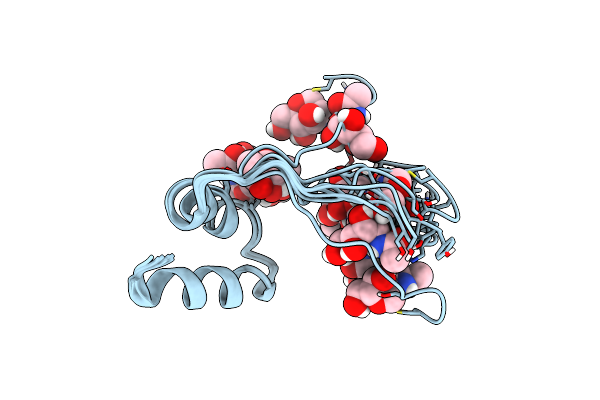 |
Nmr Shows Why A Small Chemical Change Almost Abolishes The Antimicrobial Activity Of Gccf
Organism: Lactiplantibacillus plantarum
Method: SOLUTION NMR Release Date: 2023-07-05 Classification: ANTIMICROBIAL PROTEIN Ligands: NAG |
 |
Structure Of Two-Domain Translational Regulator Yih1 Reveals A Possible Mechanism Of Action
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: SOLUTION NMR Release Date: 2021-02-10 Classification: TRANSLATION |
 |
Structure Of Two-Domain Translational Regulator Yih1 Reveals A Possible Mechanism Of Action
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: SOLUTION NMR Release Date: 2021-02-10 Classification: TRANSLATION |
 |
Structure Of Two-Domain Translational Regulator Yih1 Reveals A Possible Mechanism Of Action
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: SOLUTION NMR, SOLUTION SCATTERING Release Date: 2018-11-28 Classification: TRANSLATION |
 |
Crystal Structure Of Chorismate Mutase From Helicobacter Pylori In Complex With Prephenate
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-11-07 Classification: ISOMERASE Ligands: PRE, PYR, PHB |
 |
The Structure Of A Dimeric Type Ii Dah7Ps Associated With Pyocyanin Biosynthesis In Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-10-03 Classification: TRANSFERASE Ligands: PEP, CO, CL |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2018-02-28 Classification: TRANSFERASE Ligands: PEP, TRP, PO4, CO, CL |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-02-28 Classification: TRANSFERASE Ligands: PO4, CO, PEP, TYR, CL |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2018-02-28 Classification: TRANSFERASE Ligands: CO, PEP, CL, ACT |
 |
Organism: Neisseria meningitidis
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2015-11-25 Classification: TRANSFERASE Ligands: MN, PEG, SO4, PHE |
 |
Crystal Structure Of The Pectin Methylesterase From Aspergillus Niger In Deglycosylated Form
Organism: Aspergillus niger (strain atcc 1015 / cbs 113.46 / fgsc a1144 / lshb ac4 / nctc 3858a / nrrl 328 / usda 3528.7)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-07-01 Classification: HYDROLASE Ligands: SO4, CL, GOL, ACT |
 |
Crystal Structure Of The Pectin Methylesterase From Aspergillus Niger In Penultimately Deglycosylated Form (N-Acetylglucosamine Stub At Asn84)
Organism: Aspergillus niger (strain atcc 1015 / cbs 113.46 / fgsc a1144 / lshb ac4 / nctc 3858a / nrrl 328 / usda 3528.7)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2015-07-01 Classification: HYDROLASE Ligands: NAG, SO4, CL, GOL, ACT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2015-03-25 Classification: TRANSFERASE Ligands: SO4 |
 |
Structural Analysis Of Substrate-Mimicking Inhibitors In Complex With Neisseria Meningitidis 3 Deoxy D Arabino Heptulosonate 7 Phosphate Synthase The Importance Of Accommodating The Active Site Water
Organism: Neisseria meningitidis
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2014-10-08 Classification: TRANSFERASE Ligands: MN, GZ3 |
 |
Structural Analysis Of Substrate-Mimicking Inhibitors In Complex With Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - The Importance Of Accommodating The Active Site Water
Organism: Neisseria meningitidis
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2014-10-08 Classification: TRANSFERASE Ligands: MN, PO4, 0V5 |

