Search Count: 64
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2018-04-04 Classification: IMMUNE SYSTEM Ligands: GOL, TCE |
 |
Crystal Structure Of Beta Subunit Of Acyl-Coa Carboxylase Accd1 From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2016-04-20 Classification: LIGASE Ligands: GOL |
 |
Crystal Structure Of Human Alpha-L-Iduronidase Complex With Alpha-L-Iduronic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2015-01-14 Classification: HYDROLASE Ligands: NAG, GOL, TLA, IDR, CL, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2015-01-14 Classification: HYDROLASE Ligands: NAG, GOL, CL, SO4 |
 |
The Crystal Structure Of Beta-Hexosaminidase B In Complex With Pyrimethamine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2011-02-16 Classification: HYDROLASE Ligands: NDG, NAG, CP6, SO4 |
 |
Crystal Structure Of Probable Thiosulfate Sulfurtransferase Cysa3 (Rv3117) From Mycobacterium Tuberculosis: Monoclinic Form
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-12-08 Classification: TRANSFERASE |
 |
Crystal Structure Of Probable Thiosulfate Sulfurtransferase Cysa3 (Rv3117) From Mycobacterium Tuberculosis: Orthorhombic Form
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-12-08 Classification: TRANSFERASE Ligands: SO4, GOL |
 |
Organism: Hepatitis c virus subtype 1b
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2009-04-07 Classification: TRANSFERASE Ligands: ACY, GOL |
 |
Crystal Structure Of Haloalkane Dehalogenase Rv2579 From Mycobacterium Tuberculosis Complexed With 1,2-Dichloroethane
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2007-11-13 Classification: HYDROLASE Ligands: CL, ACT, DCE |
 |
Crystal Structure Of Haloalkane Dehalogenase Rv2579 From Mycobacterium Tuberculosis Complexed With 1,3-Propandiol
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2007-11-13 Classification: HYDROLASE Ligands: BR, PDO |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-10-23 Classification: ISOMERASE Ligands: TLA, ACY |
 |
Organism: Haemophilus influenzae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-10-23 Classification: ISOMERASE Ligands: SO4, EDO |
 |
Crystal Structure Of The Conserved Hypothetical Protein Rv1873 From Mycobacterium Tuberculosis At 1.38 A
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2007-01-30 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: SO4, GOL |
 |
Crystal Structure Of Sars Coronavirus Main Peptidase At Ph 6.0 In The Space Group P21
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2006-12-26 Classification: HYDROLASE Ligands: MES |
 |
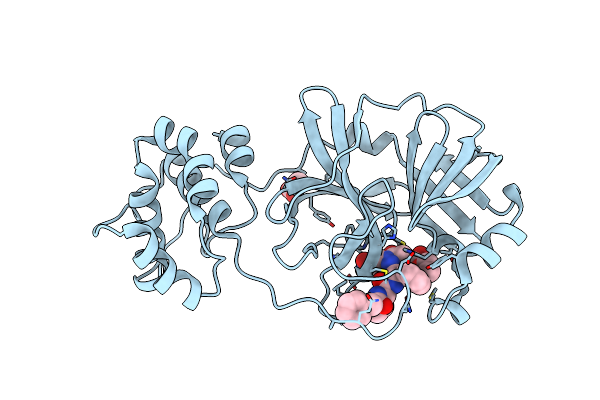
Crystal Structure Of Sars Coronavirus Main Peptidase (With An Additional Ala At The N-Terminus Of Each Protomer) In The Space Group P43212
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-12-26 Classification: HYDROLASE |
 |
Crystal Structure Of Sars Coronavirus Main Peptidase (With An Additional Ala At The N-Terminus Of Each Protomer) Inhibited By An Aza-Peptide Epoxide In The Space Group P43212
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-12-26 Classification: HYDROLASE Ligands: AZP, ACY |
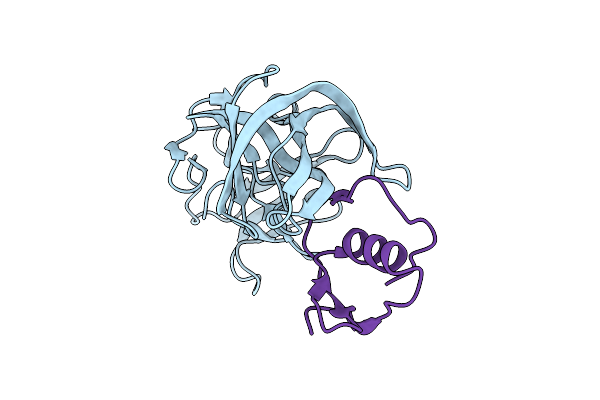 |
Molecular Structures Of The Complexes Of Sgpb With Omtky3 Aromatic P1 Variants Trp18I, His18I, Phe18I, And Tyr18I
Organism: Meleagris gallopavo, Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2006-11-21 Classification: HYDROLASE |
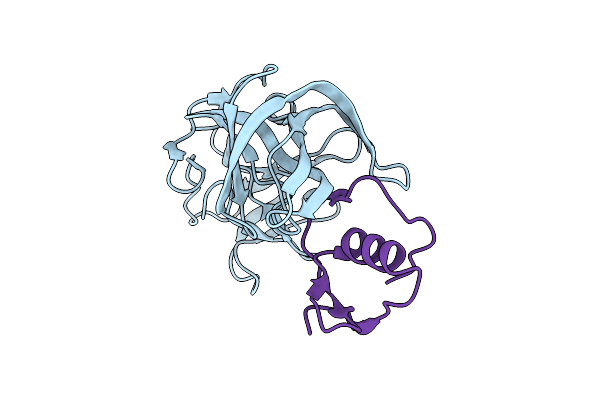 |
Molecular Structures Of The Complexes Of Sgpb With Omtky3 Aromatic P1 Variants Trp18I, His18I, Phe18I And Tyr18I
Organism: Meleagris gallopavo, Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-11-21 Classification: HYDROLASE |
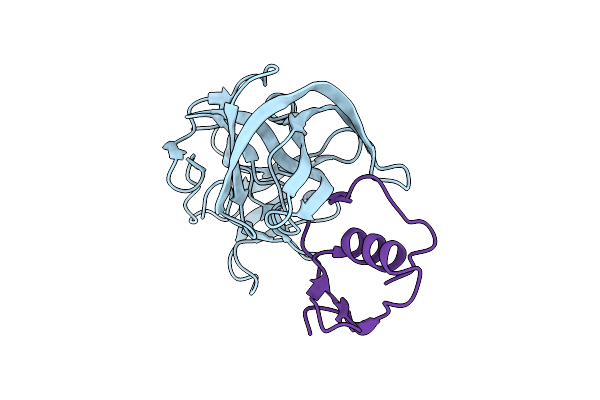 |
Accommodation Of Positively-Charged Residues In A Hydrophobic Specificity Pocket: Crystal Structures Of Sgpb In Complex With Omtky3 Variants Lys18I And Arg18I
Organism: Meleagris gallopavo, Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2006-11-21 Classification: HYDROLASE |
 |
Accommodation Of Positively-Charged Residues In A Hydrophobic Specificity Pocket: Crystal Structures Of Sgpb In Complex With Omtky3 Variants Lys18I And Arg18I
Organism: Meleagris gallopavo, Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-11-21 Classification: HYDROLASE |

