Search Count: 21
 |
Crystal Structure Of Trk-A In Complex With The Pan-Trk Kinase Inhibitor, Compound 10B.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2018-07-11 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: FKY |
 |
Crystal Structure Of Trk-A In Complex With The Pan-Trk Kinase Inhibitor, Compound 13B.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2018-07-11 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 22L, GO7 |
 |
Crystal Structure Of Trk-A In Complex With The Pan-Trk Kinase Inhibitor, Compound 19.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2018-07-11 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 22L, GOD |
 |
Crystal Structure Of Trk-A In Complex With The Pan-Trk Kinase Inhibitor, Compound 3.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2018-07-11 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: GXA |
 |
4,5 Diaryl Isoxazole Hsp90 Chaperone Inhibitors: Potential Therapeutic Agents For The Treatment Of Cancer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-12-11 Classification: CHAPERONE Ligands: 2GJ |
 |
4,5 Diaryl Isoxazole Hsp90 Chaperone Inhibitors: Potential Therapeutic Agents For The Treatment Of Cancer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-12-11 Classification: CHAPERONE Ligands: 2EQ |
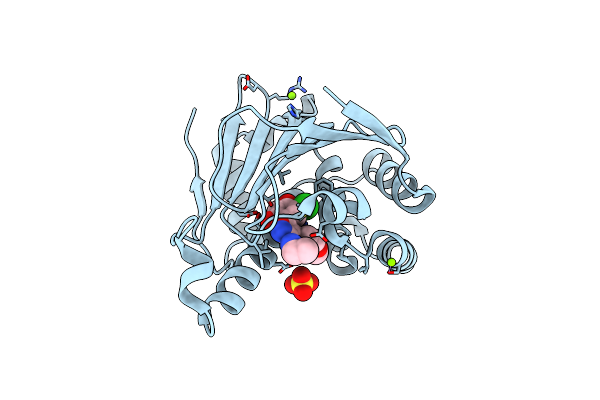 |
Inhibition Of The Hsp90 Molecular Chaperone In Vitro And In Vivo By Novel, Synthetic, Potent Resorcinylic Pyrazole, Isoxazole Amide Analogs
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-04-24 Classification: CHAPERONE Ligands: MG, SO4, 2GG |
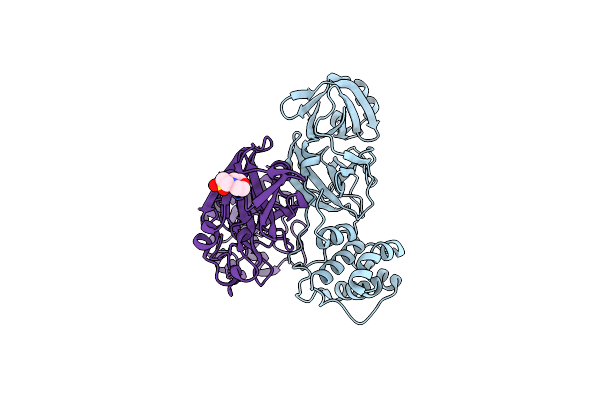 |
Crystal Structure Of Sars Coronavirus Main Peptidase At Ph 6.0 In The Space Group P21
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2006-12-26 Classification: HYDROLASE Ligands: MES |
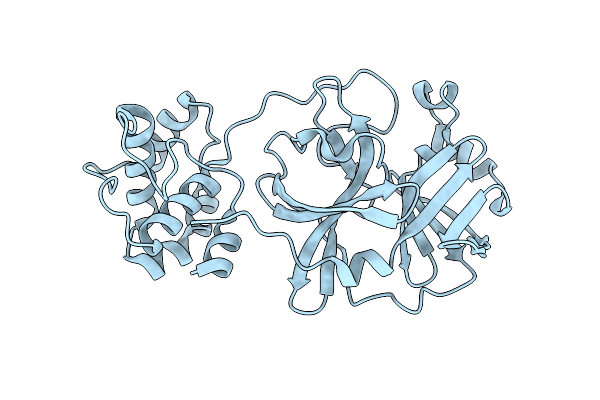 |
Crystal Structure Of Sars Coronavirus Main Peptidase (With An Additional Ala At The N-Terminus Of Each Protomer) In The Space Group P43212
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-12-26 Classification: HYDROLASE |
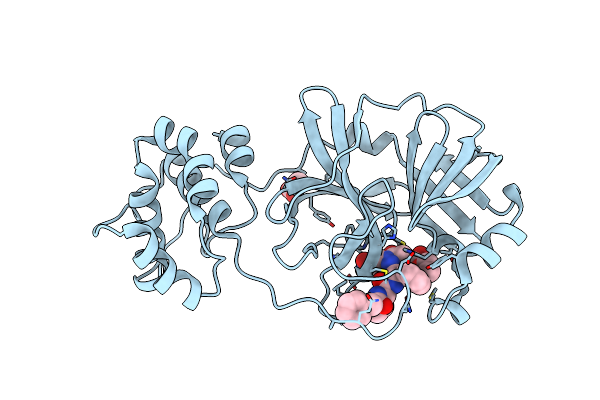 |
Crystal Structure Of Sars Coronavirus Main Peptidase (With An Additional Ala At The N-Terminus Of Each Protomer) Inhibited By An Aza-Peptide Epoxide In The Space Group P43212
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-12-26 Classification: HYDROLASE Ligands: AZP, ACY |
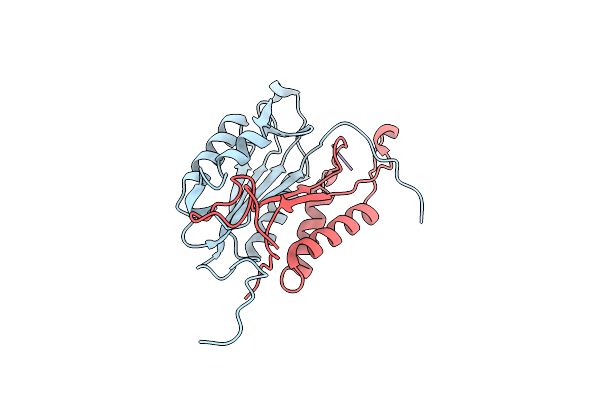 |
Crystal Structures Of Caspase-3 In Complex With Aza-Peptide Michael Acceptor Inhibitors.
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2006-09-20 Classification: HYDROLASE/HYDROLASE INHIBITOR |
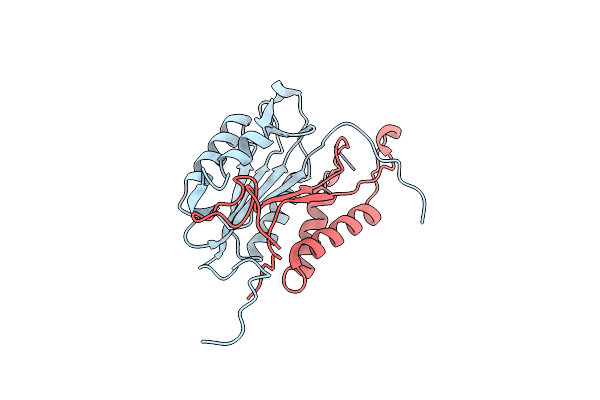 |
Crystal Structures Of Caspase-3 In Complex With Aza-Peptide Michael Acceptor Inhibitors.
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2006-09-20 Classification: HYDROLASE/HYDROLASE INHIBITOR |
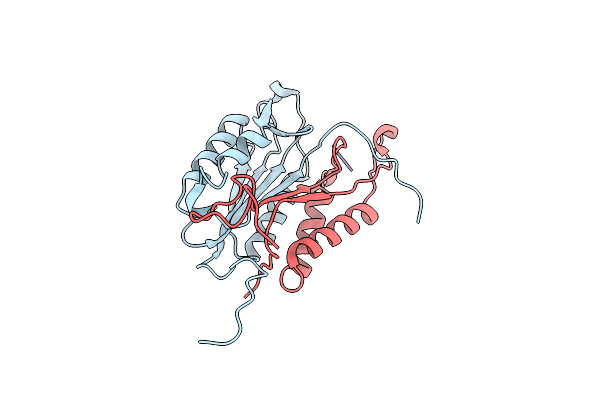 |
Crystal Structures Of Caspase-3 In Complex With Aza-Peptide Michael Acceptor Inhibitors.
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2006-09-20 Classification: HYDROLASE/HYDROLASE INHIBITOR |
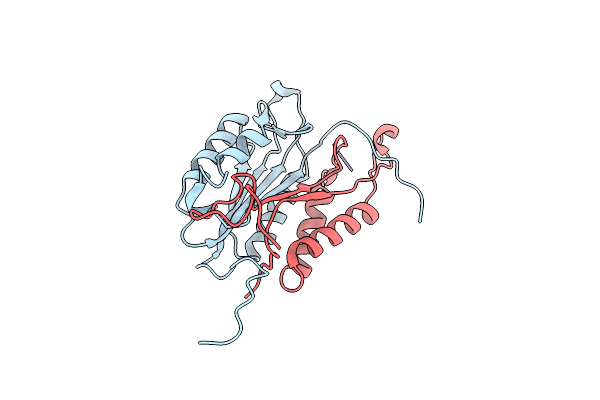 |
Crystal Structures Of Caspase-3 In Complex With Aza-Peptide Michael Acceptor Inhibitors.
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2006-09-20 Classification: HYDROLASE/HYDROLASE INHIBITOR |
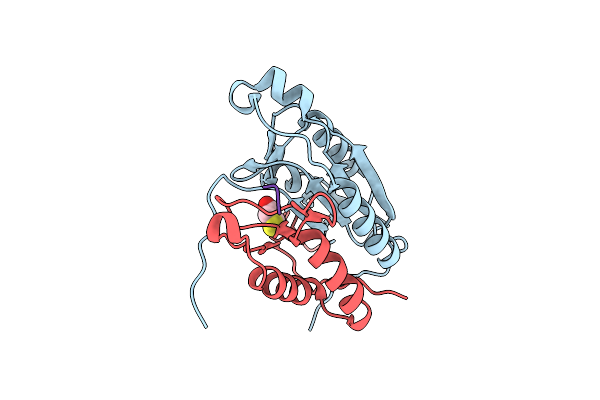 |
Crystal Structure Of Caspase-8 In Complex With Aza-Peptide Michael Acceptor Inhibitor
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2006-09-20 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: DTD |
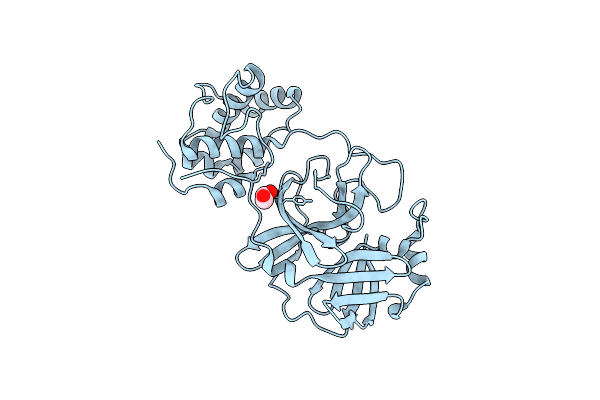 |
Crystal Structure Of Unbound Sars Coronavirus Main Peptidase In The Space Group C2
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2005-10-25 Classification: HYDROLASE Ligands: CL, EDO |
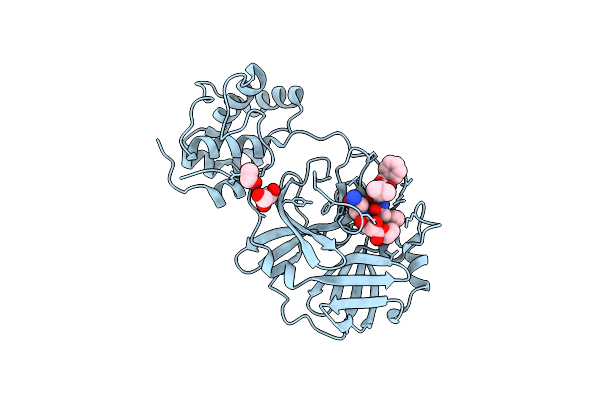 |
Crystal Structures Of Sars Coronavirus Main Peptidase Inhibited By An Aza-Peptide Epoxide In The Space Group C2
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2005-10-25 Classification: HYDROLASE Ligands: AZP, EDO, GOL |
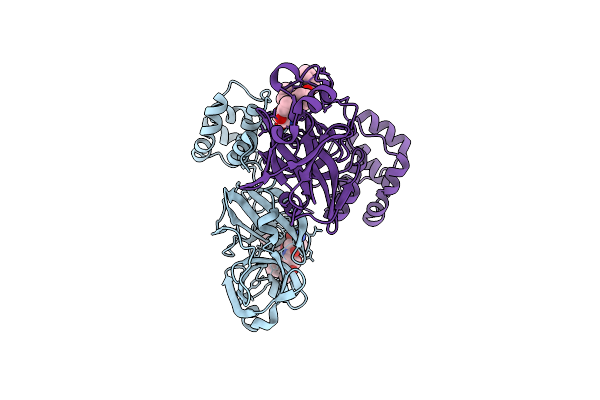 |
Crystal Structures Of Sars Coronavirus Main Peptidase Inhibited By An Aza-Peptide Epoxide In Space Group P212121
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2005-10-25 Classification: HYDROLASE Ligands: AZP |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2005-09-29 Classification: CHAPERONE Ligands: CT5 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2005-09-29 Classification: CHAPERONE Ligands: KJ2 |

