Search Count: 31
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: MEMBRANE PROTEIN Ligands: HEB, NDP, FAD, PX2, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: MEMBRANE PROTEIN Ligands: HEB, NDP, FAD, D12, D10, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: MEMBRANE PROTEIN Ligands: HEB, NDP, FAD, ZN, D12 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: MEMBRANE PROTEIN Ligands: FAD, D12, D10, NAP, HEB, ZN |
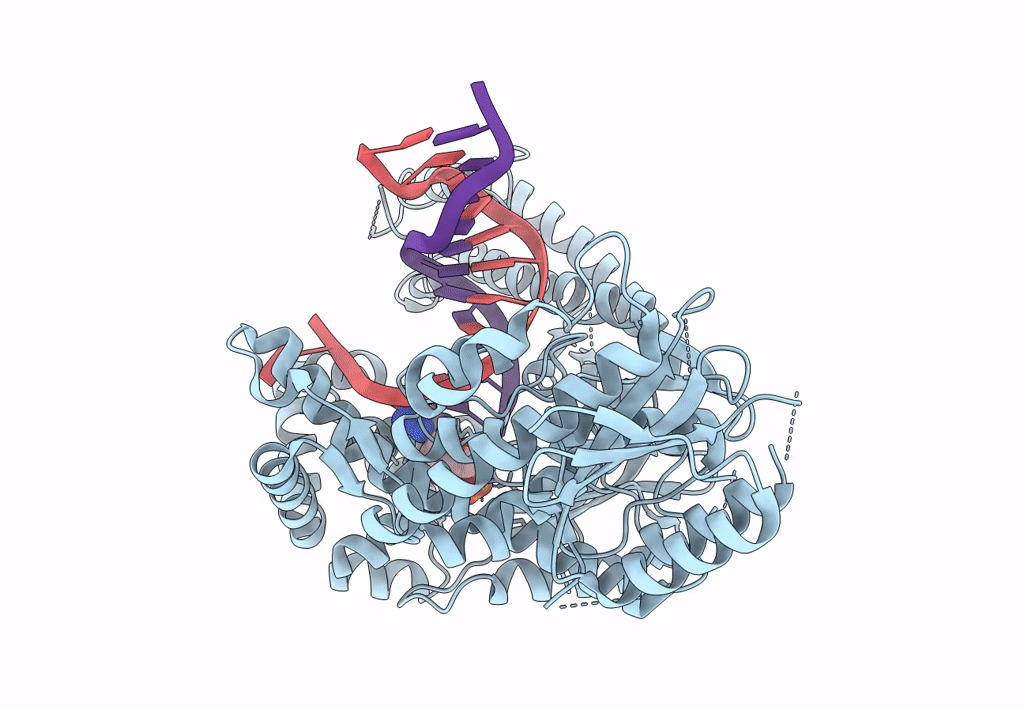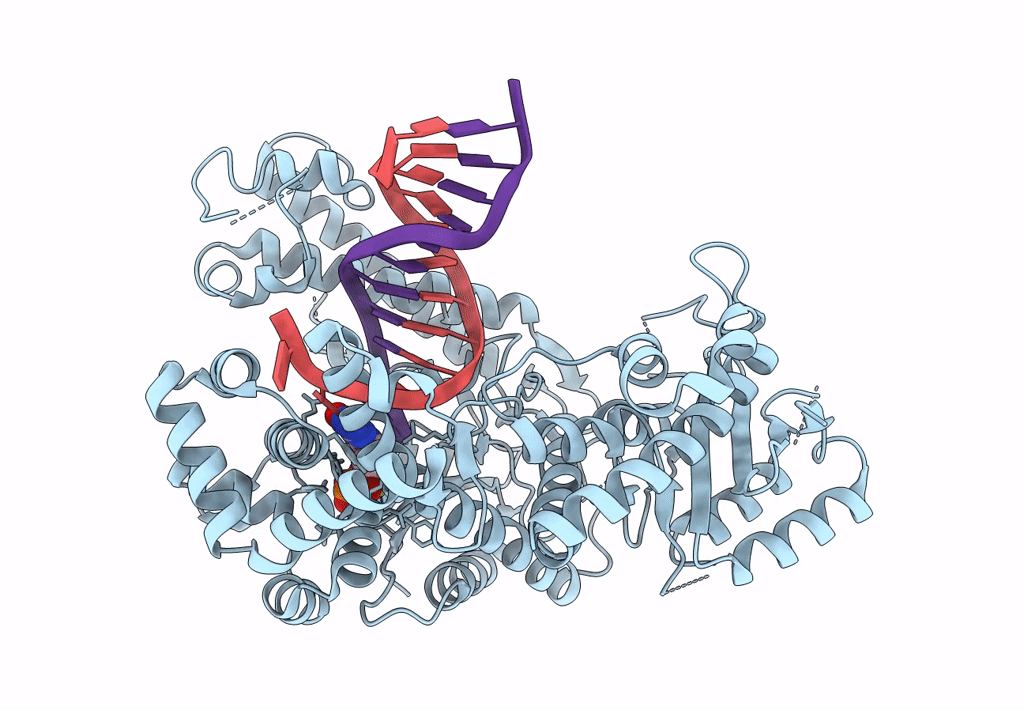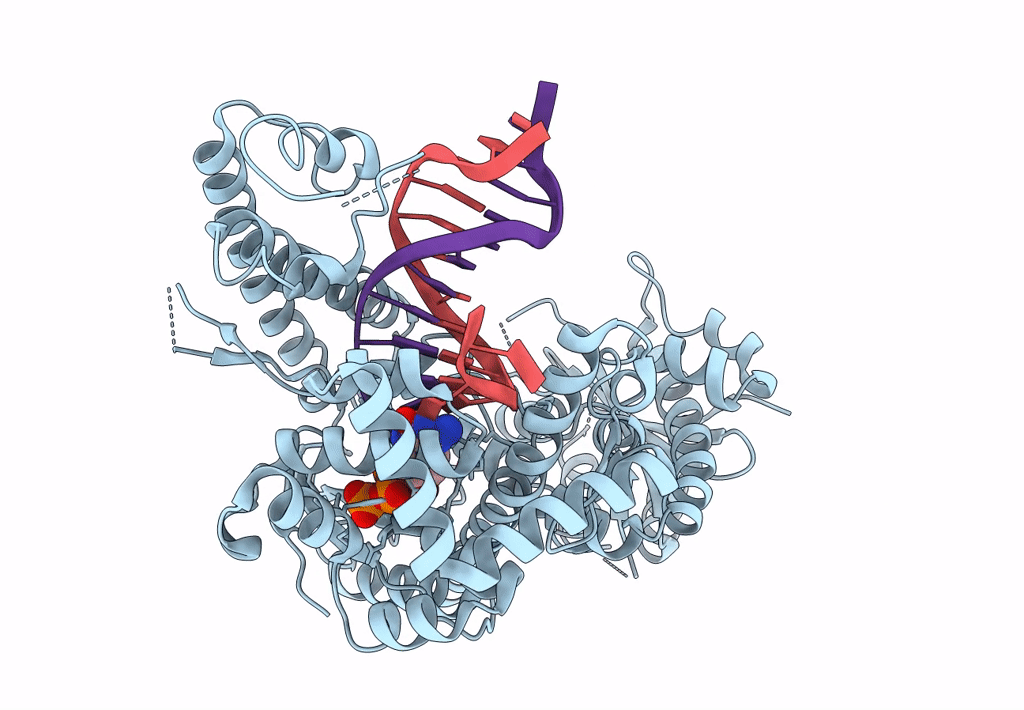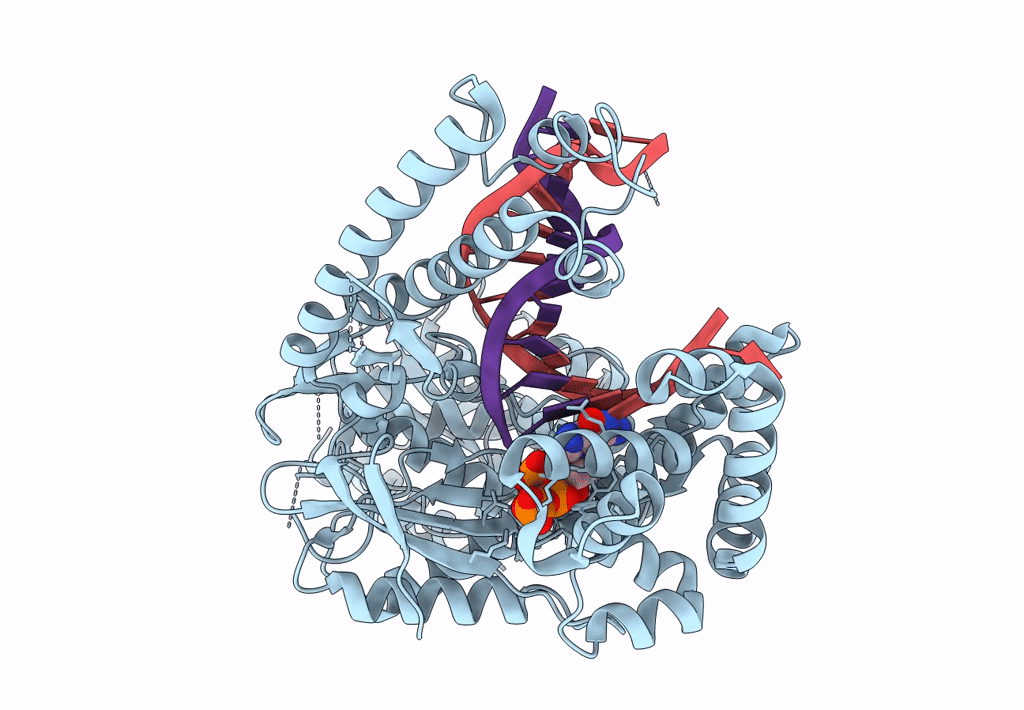 |
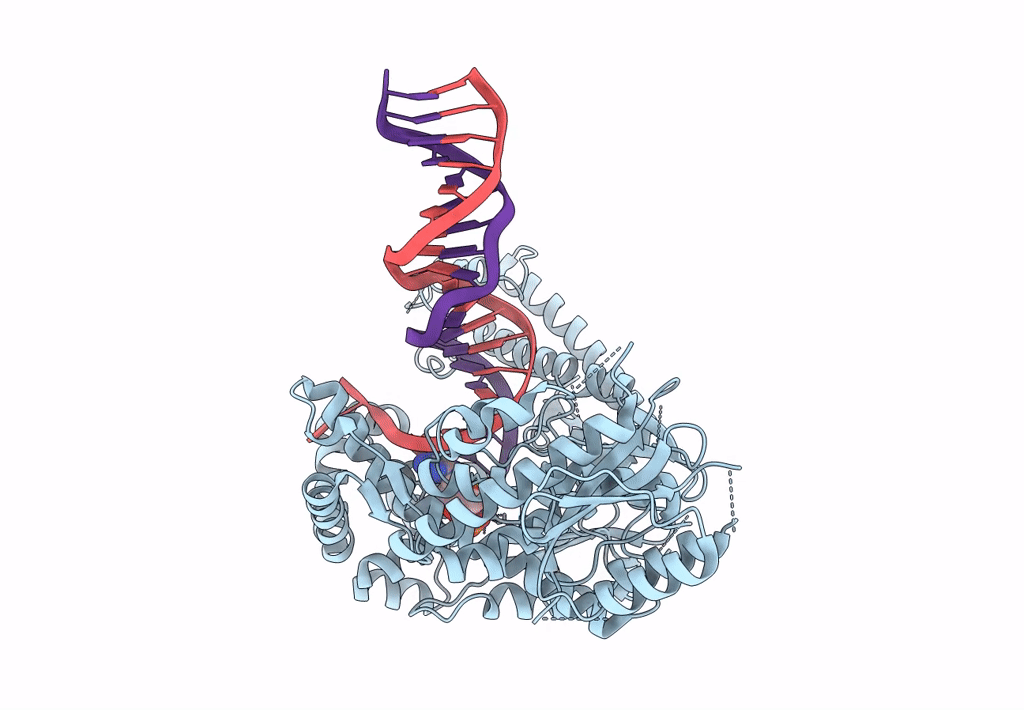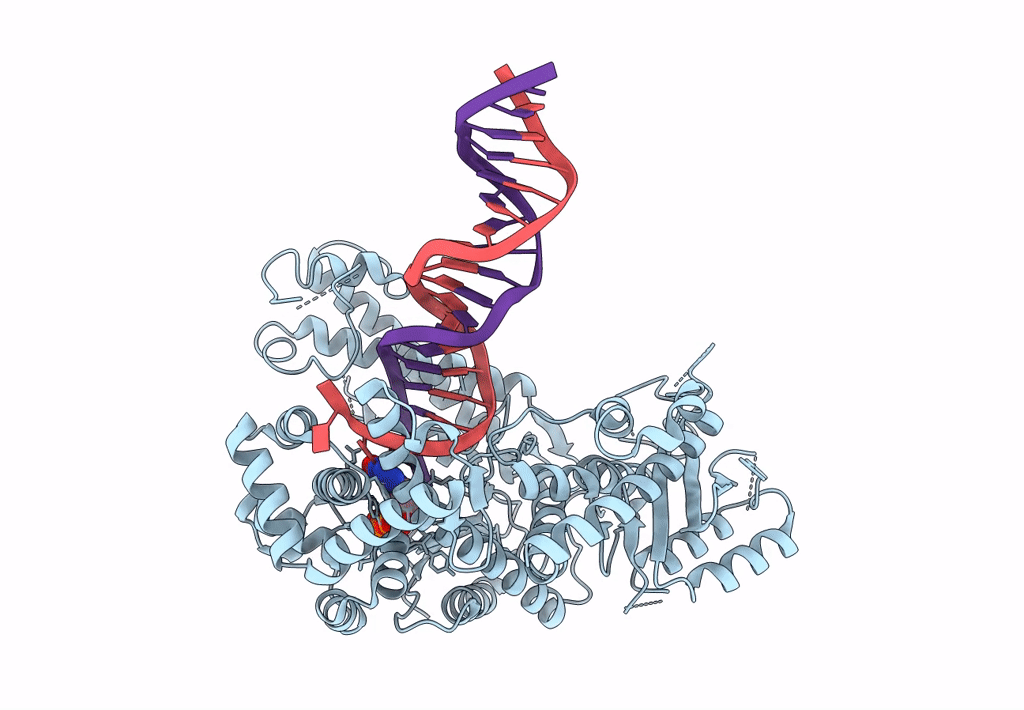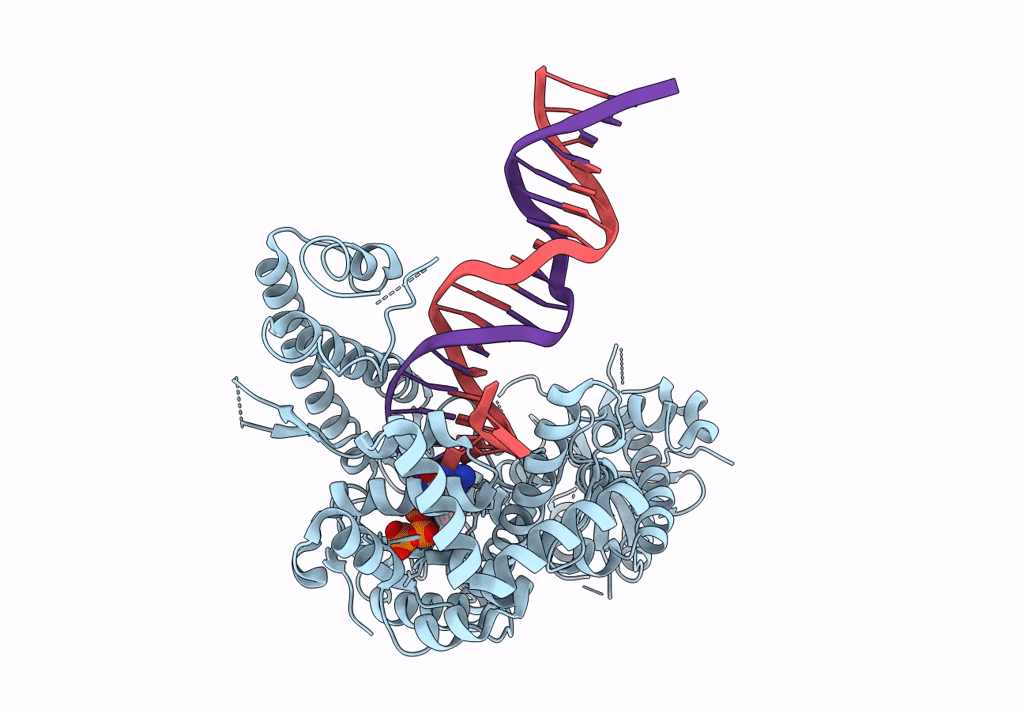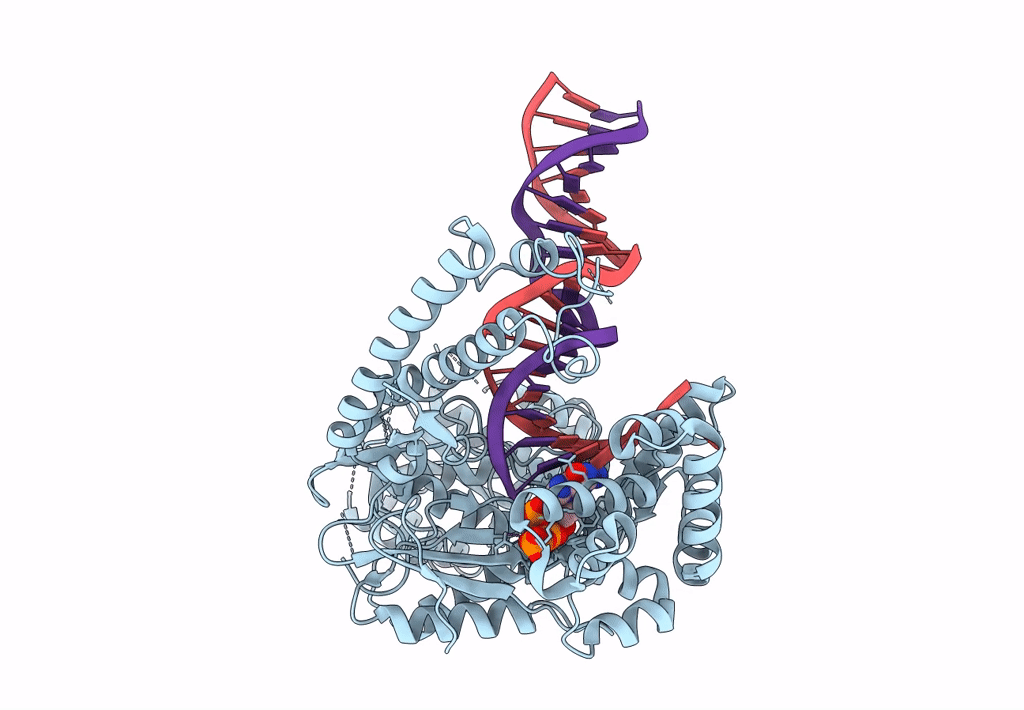
Structure Of Lates Calcarifer Dna Polymerase Theta Polymerase Domain With Long Duplex Dna, Complex Ia
Organism: Lates calcarifer
Method: ELECTRON MICROSCOPY Release Date: 2022-12-14 Classification: DNA BINDING PROTEIN/DNA Ligands: DGT, MG |
 |
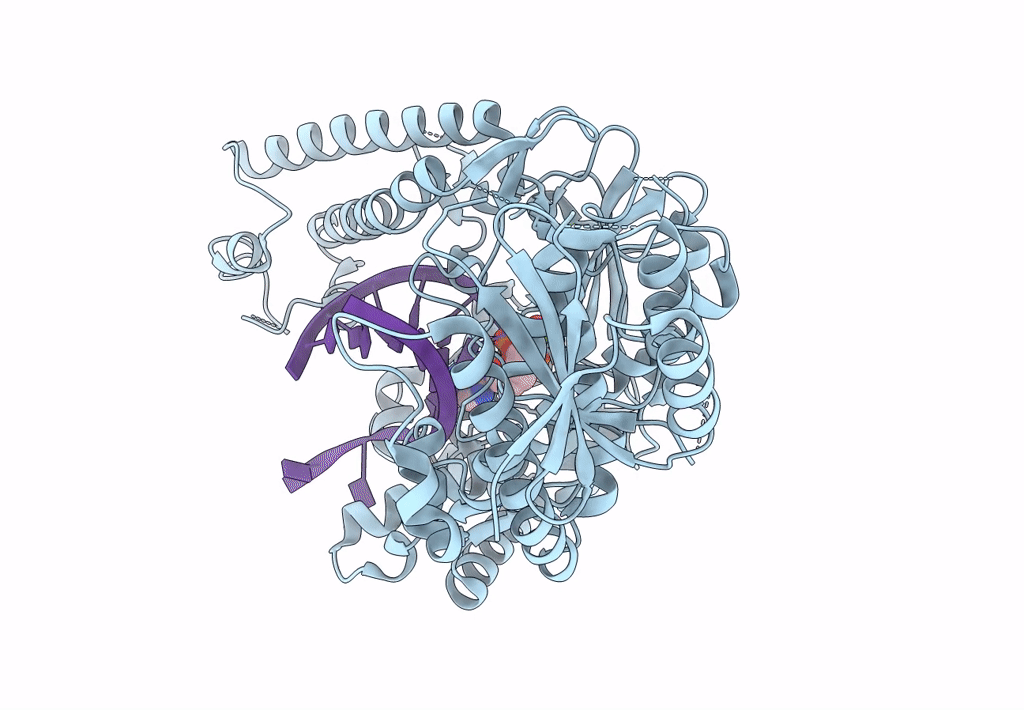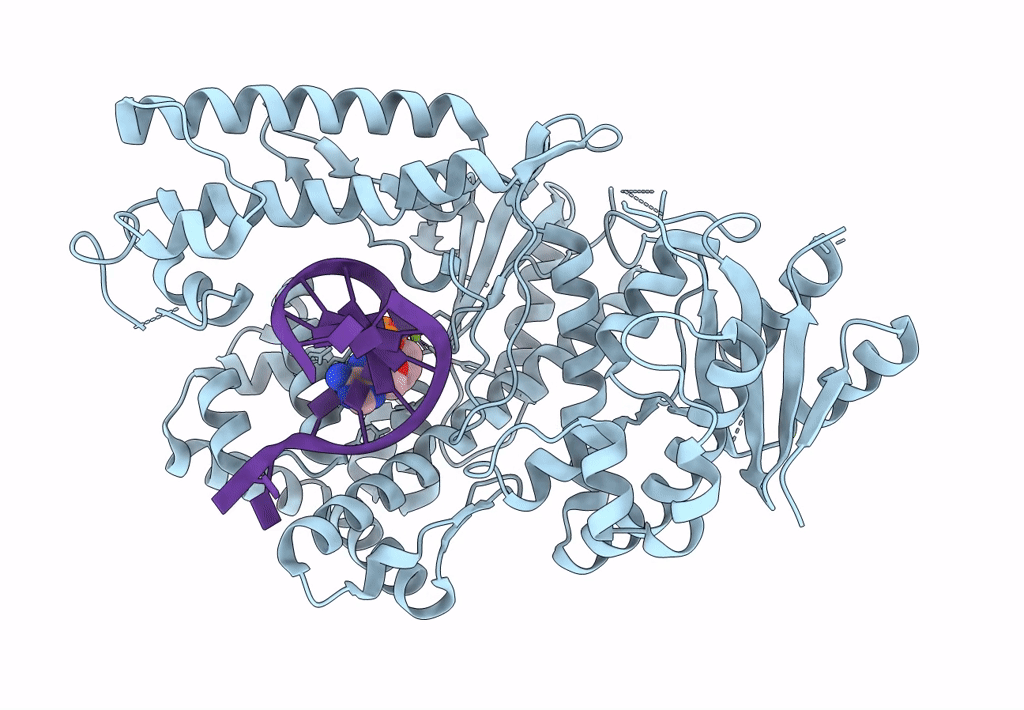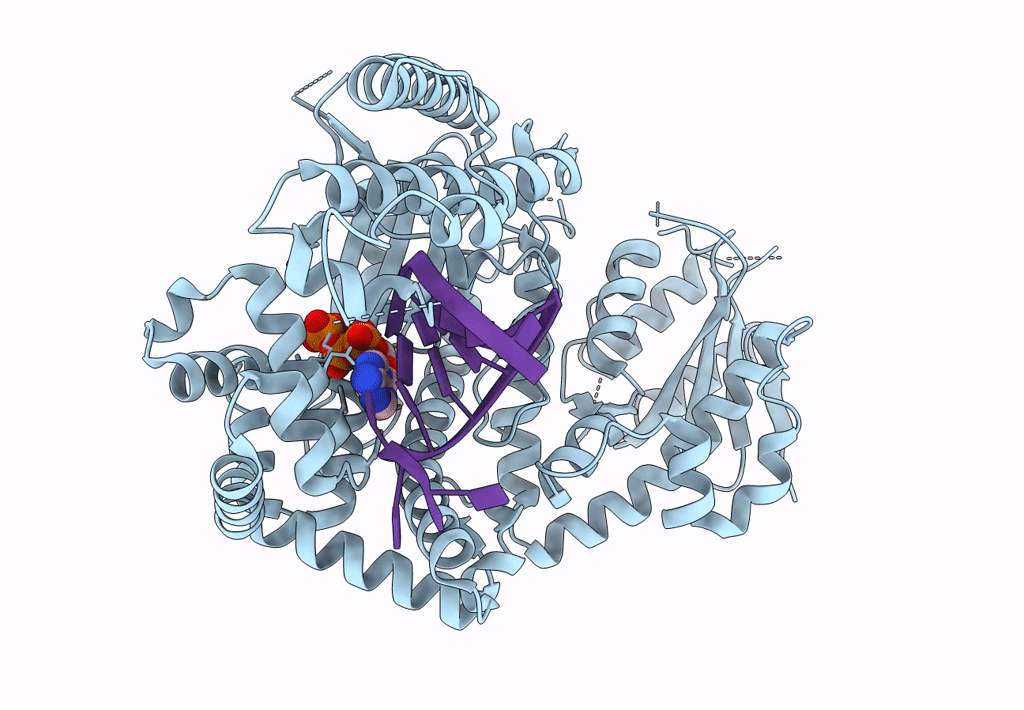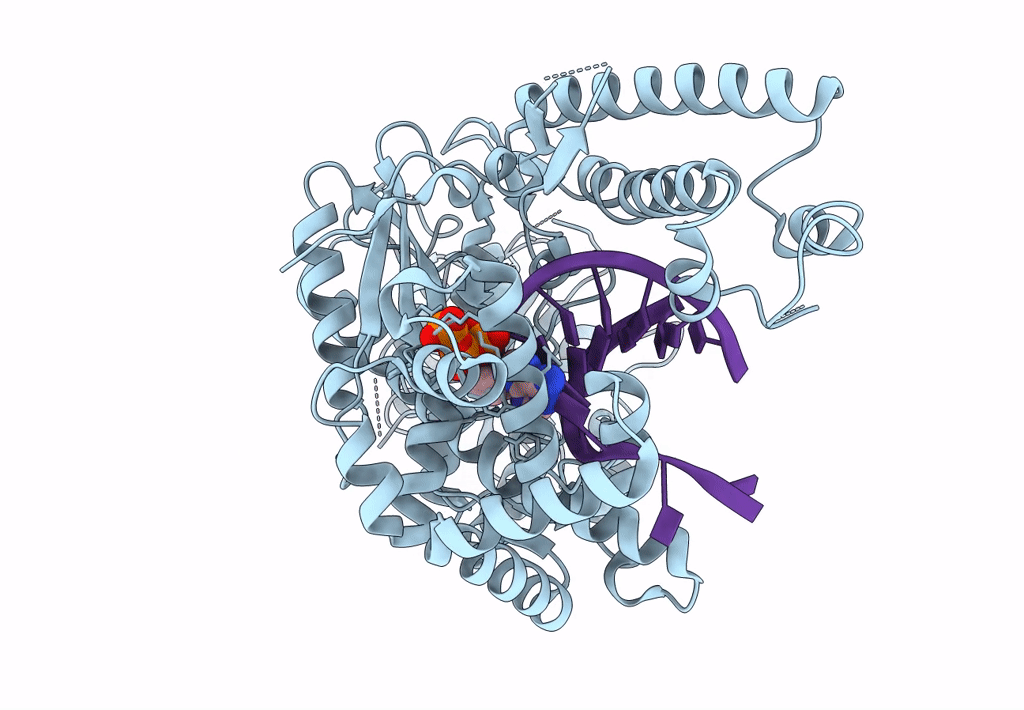
Structure Of Lates Calcarifer Dna Polymerase Theta Polymerase Domain With Long Duplex Dna, Complex Ia
Organism: Lates calcarifer
Method: ELECTRON MICROSCOPY Release Date: 2022-12-14 Classification: DNA BINDING PROTEIN Ligands: DGT, MG |
 |
Structure Of Lates Calcarifer Dna Polymerase Theta Polymerase Domain With Hairpin Dna
Organism: Lates calcarifer
Method: ELECTRON MICROSCOPY Release Date: 2022-12-14 Classification: DNA BINDING PROTEIN/DNA Ligands: MG, DDS |
 |
Encoded Conformational Dynamics Of The Hiv Splice Site A3 Regulatory Locus: Implications For Differential Binding Of Hnrnp Splicing Auxiliary Factors
Organism: Hiv whole-genome vector aa1305#18
Method: SOLUTION NMR, SOLUTION SCATTERING Release Date: 2022-06-29 Classification: RNA |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: RIBOSOME |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: RIBOSOME |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-06-30 Classification: HYDROLASE Ligands: ZN, DMS, GLY, CL, BTB |
 |
Glycoside Hydrolase Family 16 Endo-Glucanase From Bacteroides Ovatus In Complex With G4G3G-2F-Dnp
Organism: Bacteroides ovatus
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2021-05-19 Classification: HYDROLASE |
 |
Glycoside Hydrolase Family 16 Endo-Glucanase From Bacteroides Ovatus In Complex With G4G4G3G-Nhcoch2Br
Organism: Bacteroides ovatus (strain atcc 8483 / dsm 1896 / jcm 5824 / nctc 11153)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-01-13 Classification: HYDROLASE |
 |
Bacteroides Uniformis Endo-Laminarinase Bugh158 From The Beta(1,3)-Glucan Utilization Locus
Organism: Bacteroides uniformis
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2020-04-01 Classification: HYDROLASE Ligands: ACT, SO4 |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2019-09-18 Classification: RIBOSOME |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2019-09-18 Classification: RIBOSOME Ligands: GNP |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2019-09-18 Classification: RIBOSOME |
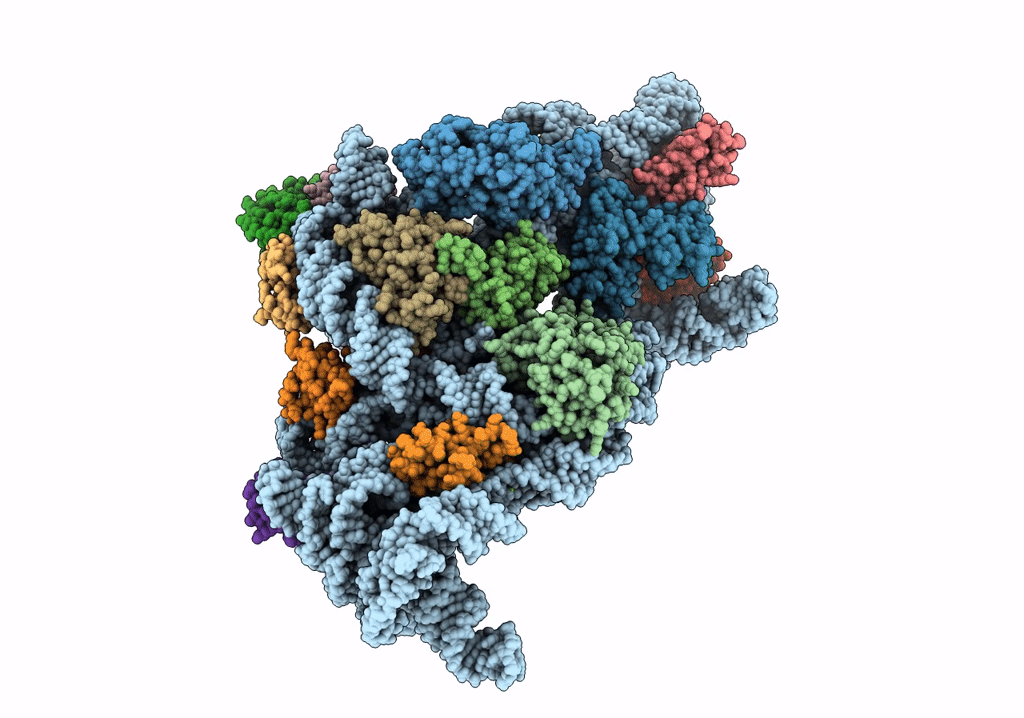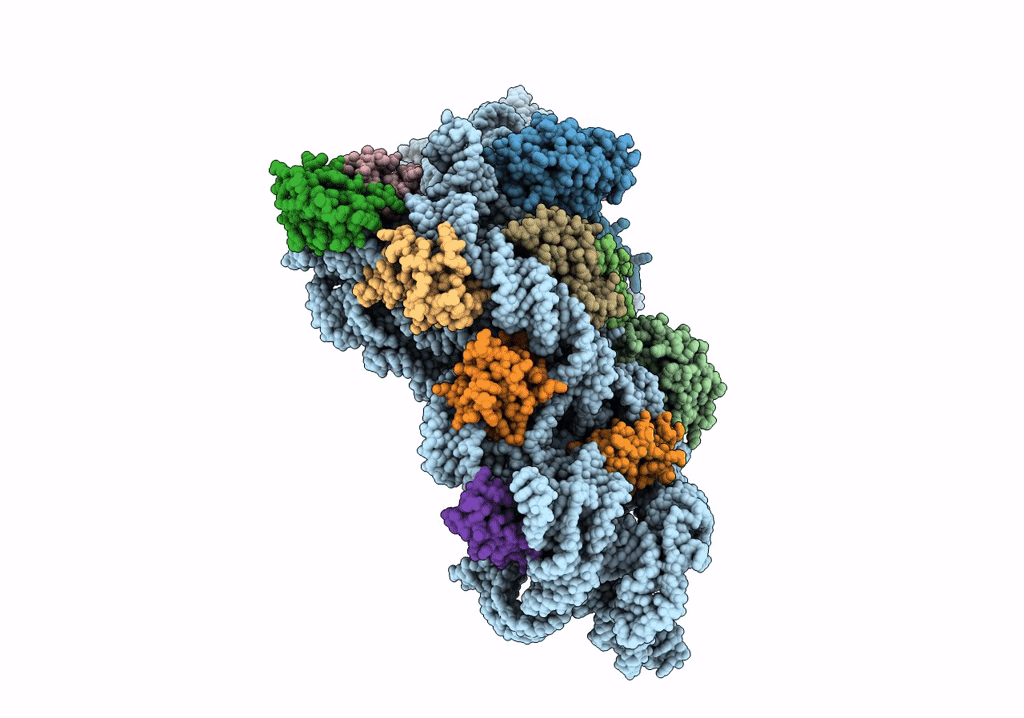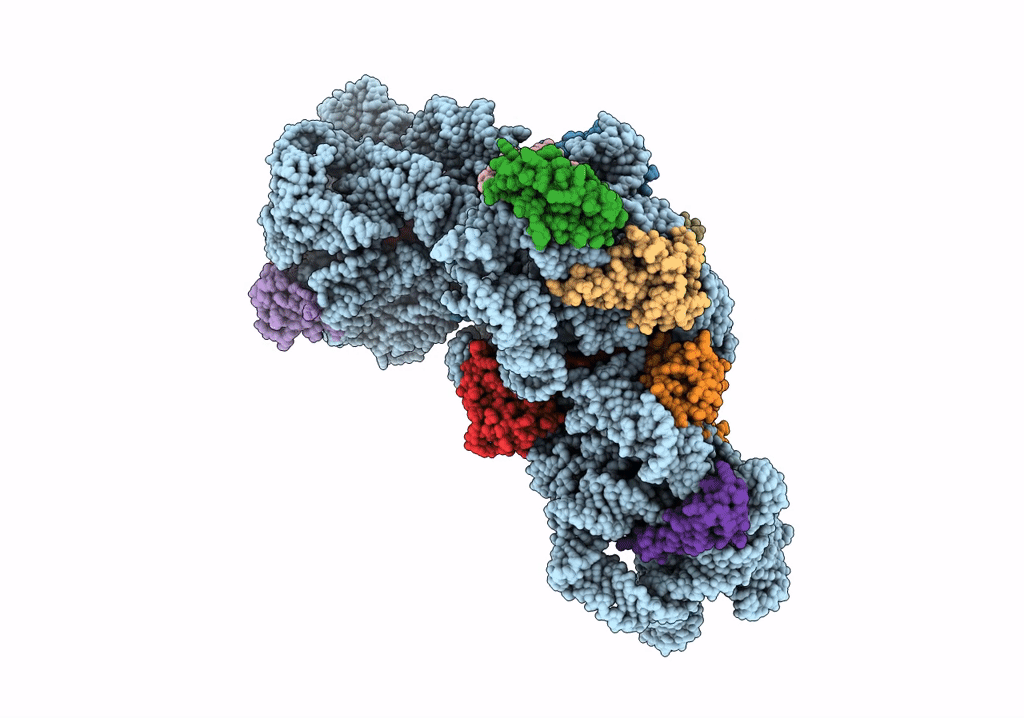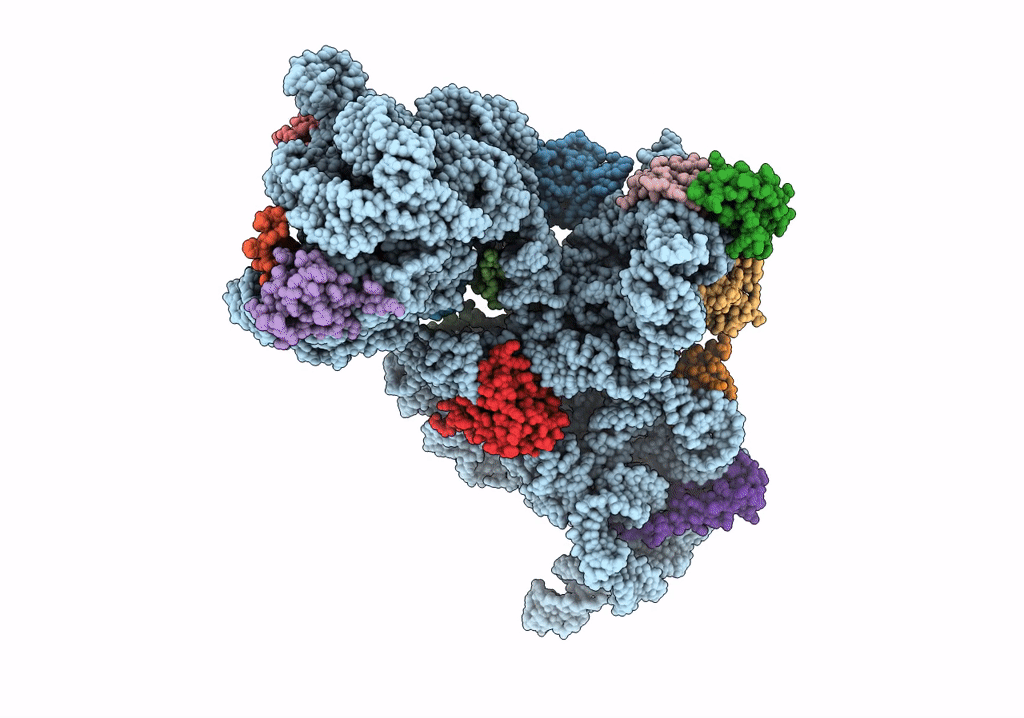 |
Organism: Escherichia coli, Escherichia coli h736
Method: ELECTRON MICROSCOPY Release Date: 2019-06-26 Classification: RIBOSOME Ligands: MG |
 |
Structure Of An Endo-Xyloglucanase From Cellvibrio Japonicus Complexed With Xxxg(2F)-Beta-Dnp
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-10-03 Classification: HYDROLASE Ligands: SO4, GOL, 1PE |
 |
Structure Of A Covalent Complex Of Endo-Xyloglucanase From Cellvibrio Japonicus After Reacting With Xxxg(2F)-Beta-Dnp
Organism: Cellvibrio japonicus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-10-03 Classification: HYDROLASE Ligands: GOL, SO4, PGE, MES, 1PE |

