Search Count: 21
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-02-05 Classification: DNA BINDING PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2020-02-05 Classification: DNA BINDING PROTEIN |
 |
Organism: Hepatitis c virus isolate hc-j4
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2014-12-31 Classification: TRANSFERASE |
 |
Organism: Hepatitis c virus isolate hc-j4
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2014-12-31 Classification: TRANSFERASE Ligands: MN, UTP |
 |
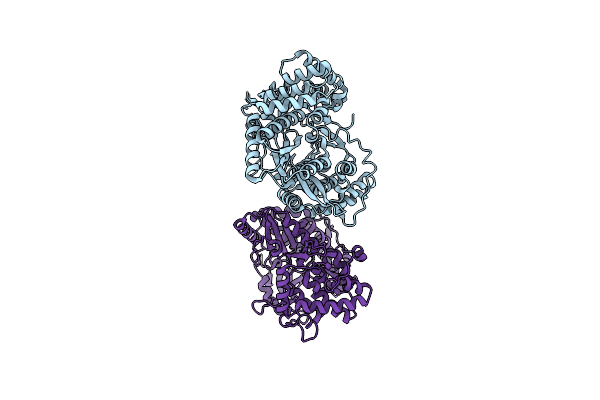
Organism: Hepatitis c virus isolate hc-j4
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2014-12-31 Classification: TRANSFERASE |
 |
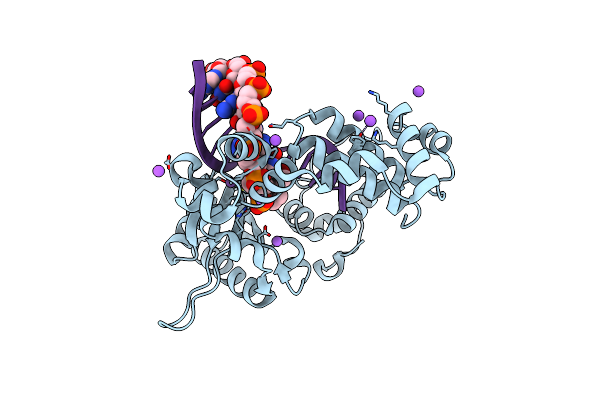
Organism: Hepatitis c virus isolate hc-j4
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2014-12-31 Classification: TRANSFERASE |
 |
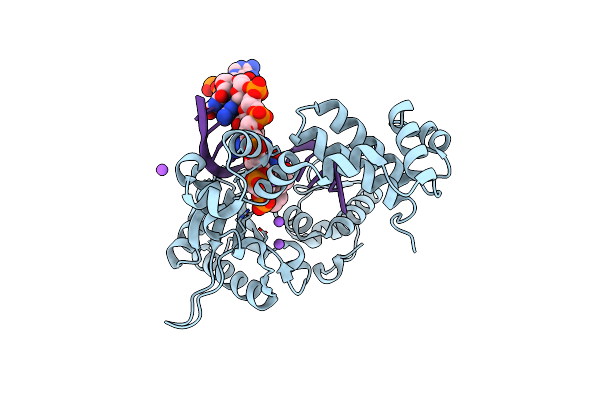
Co-Crystal Structure Of Wild Type Rat Polymerase Beta: Enzyme-Dna Binary Complex
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2013-01-16 Classification: Transferase/DNA |
 |
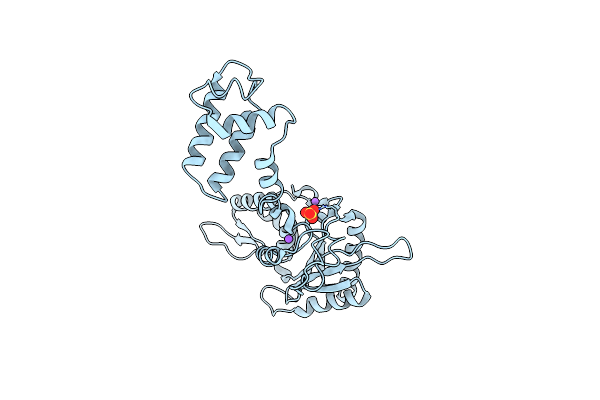
Co-Crystal Structure Of K72E Variant Of Rat Polymerase Beta: Enzyme-Dna Binary Complex
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2013-01-16 Classification: Transferase/DNA |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2013-01-16 Classification: TRANSFERASE |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-12-05 Classification: TRANSFERASE |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-12-05 Classification: TRANSFERASE, LYASE |
 |
Co-Crystal Structure Of Rat Dna Polymerase Beta Mutator I260Q: Enzyme-Dna-Ddttp
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2012-12-05 Classification: TRANSFERASE, LYASE/DNA Ligands: D3T, NA |
 |
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:3.71 Å Release Date: 2012-10-03 Classification: APOPTOSIS |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2012-07-04 Classification: DNA BINDING PROTEIN/DNA |
 |
Fidelity Properties And Structure Of M282L Mutator Mutant Of Dna Polymerase: Subtle Structural Changes Influence The Mechanism Of Nucleotide Discrimination
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2001-08-03 Classification: TRANSFERASE |
 |
The Active Site Base Controls Cofactor Reactivity In Escherichia Coli Amine Oxidase: X-Ray Crystallographic Studies With Mutational Variants.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2000-02-29 Classification: OXIDOREDUCTASE Ligands: CU, CA |
 |
The Active Site Base Controls Cofactor Reactivity In Escherichia Coli Amine Oxidase : X-Ray Crystallographic Studies With Mutational Variants
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1999-08-24 Classification: OXIDOREDUCTASE Ligands: CU, CA |
 |
The Active Site Base Controls Cofactor Reactivity In Escherichia Coli Amine Oxidase : X-Ray Crystallographic Studies With Mutational Variants
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1999-08-24 Classification: OXIDOREDUCTASE Ligands: CU, CA |
 |
The Active Site Base Controls Cofactor Reactivity In Escherichia Coli Amine Oxidase : X-Ray Crystallographic Studies With Mutational Variants
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1999-08-23 Classification: OXIDOREDUCTASE Ligands: CU, CA, GOL |
 |
Human Immunodeficiency Virus Type 1 Reverse Transcriptase Complexed With A 33-Base Nucleotide Rna Pseudoknot
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:4.75 Å Release Date: 1999-01-13 Classification: TRANSFERASE/RNA |

