Search Count: 19
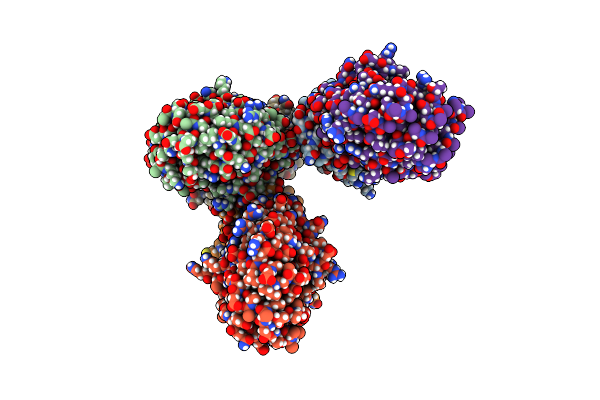 |
Crystal Structure Of Archaeoglobus Fulgidus (S)-3-O-Geranylgeranylglyceryl Phosphate Synthase
Organism: Archaeoglobus fulgidus dsm 4304
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2024-10-16 Classification: TRANSFERASE Ligands: PO4, SO4 |
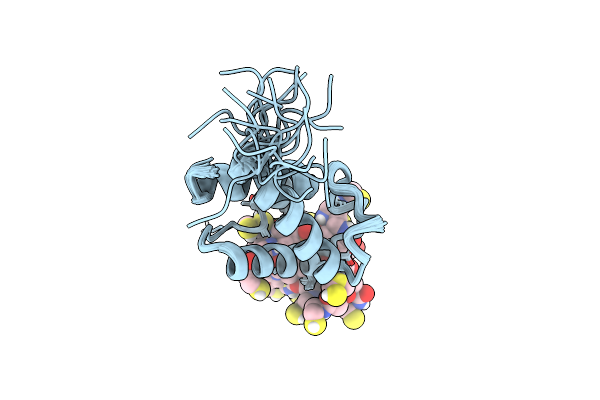 |
Organism: Streptomyces virginiae
Method: SOLUTION NMR Release Date: 2023-03-22 Classification: TRANSFERASE Ligands: PNS |
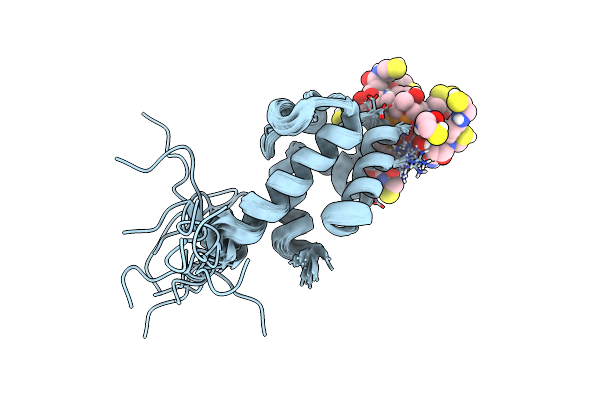 |
Organism: Streptomyces virginiae
Method: SOLUTION NMR Release Date: 2023-03-22 Classification: TRANSFERASE Ligands: PNS |
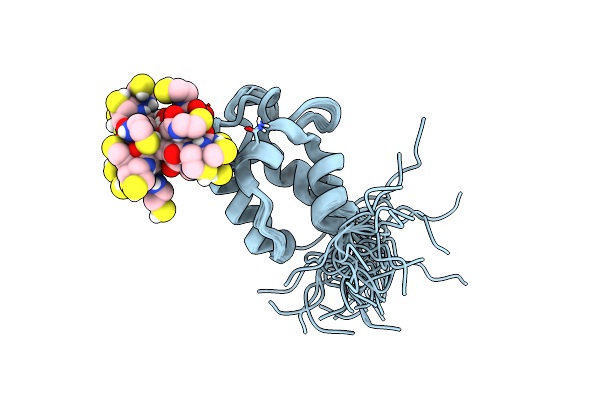 |
Organism: Streptomyces virginiae
Method: SOLUTION NMR Release Date: 2023-03-22 Classification: TRANSFERASE Ligands: PNS |
 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-03-15 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, CL, NA, PNS |
 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-03-15 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, IMD, CL |
 |
Organism: Ilumatobacter coccineus ym16-304
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-08-21 Classification: LYASE |
 |
Organism: Ilumatobacter coccineus ym16-304
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-08-21 Classification: LYASE |
 |
Organism: Ilumatobacter coccineus ym16-304
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-08-21 Classification: LYASE Ligands: BU3, CL |
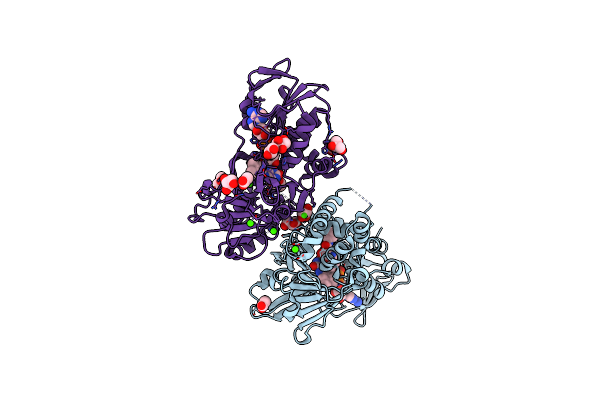 |
Crystal Structure Of A Dual Function Amine Oxidase/Cyclase In Complex With Substrate Analogues
Organism: Streptomyces rochei
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-09-19 Classification: FLAVOPROTEIN Ligands: FAD, ACT, CJ8, CA, GOL, CJE |
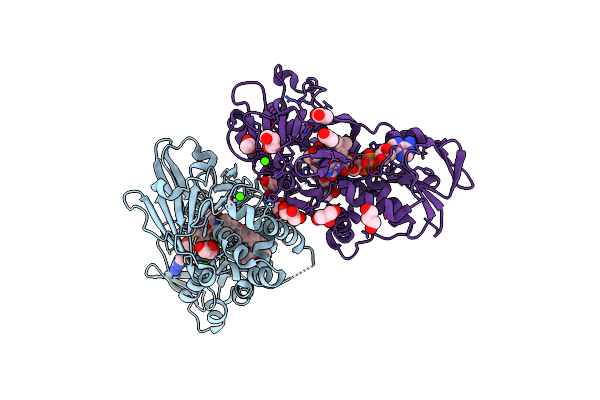 |
Organism: Streptomyces rochei
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-09-19 Classification: FLAVOPROTEIN Ligands: FAD, CA, CWH, GOL, ACT |
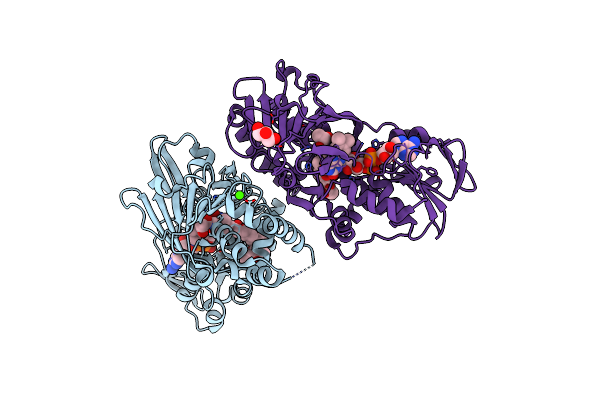 |
Organism: Streptomyces rochei subsp. volubilis
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2018-09-19 Classification: FLAVOPROTEIN Ligands: FAD, CA, CWH, GOL |
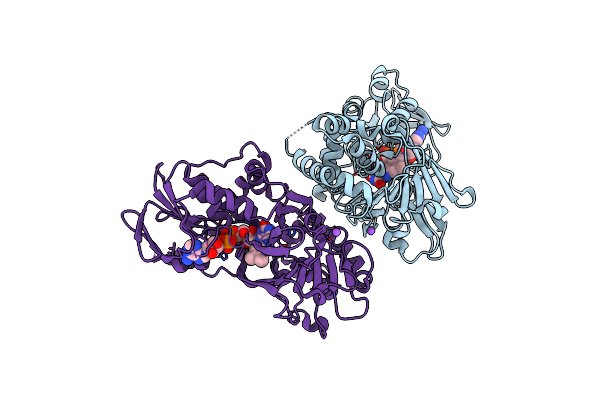 |
Organism: Streptomyces rochei subsp. volubilis
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2018-09-19 Classification: FLAVOPROTEIN Ligands: FAD, OXY, NA |
 |
Organism: Streptomyces virginiae
Method: SOLUTION NMR Release Date: 2016-03-23 Classification: PROTEIN BINDING |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2012-05-23 Classification: CELL CYCLE/INHIBITOR Ligands: GDP, 9PC |
 |
Structural Features Of The Complex Between The Dsbd N-Terminal And The Pilb N-Terminal Domains From Neisseria Meningitidis
Organism: Neisseria meningitidis serogroup a, Neisseria meningitidis serogroup b
Method: SOLUTION NMR Release Date: 2009-05-19 Classification: OXIDOREDUCTASE |
 |
Solution Structure Of The C103S Mutant Of The N-Terminal Domain Of Dsbd From Neisseria Meningitidis
Organism: Neisseria meningitidis serogroup b
Method: SOLUTION NMR Release Date: 2008-11-11 Classification: OXIDOREDUCTASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-06-06 Classification: HYDROLASE Ligands: INZ |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2001-06-06 Classification: HYDROLASE Ligands: INX |

