Search Count: 44
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: TRANSPORT PROTEIN Ligands: GOL, SO4, EDO |
 |
Crystal Structure Of Human Vps29 Complexed With Rapid-Derived Cyclic Peptide Rt-L1
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2021-07-21 Classification: PROTEIN TRANSPORT/INHIBITOR Ligands: MLI, GOL, O4B |
 |
Crystal Structure Of Human Vps29 Complexed With Rapid-Derived Cyclic Peptide Rt-D1
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2021-07-14 Classification: PROTEIN TRANSPORT/INHIBITOR Ligands: GOL, FMT |
 |
Crystal Structure Of Human Vps29 Complexed With Rapid-Derived Cyclic Peptide Rt-D2
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2021-07-14 Classification: PROTEIN TRANSPORT/INHIBITOR Ligands: GOL, FMT |
 |
Crystal Structure Of Chaetomium Thermophilum Vps29 Complexed With Rapid-Derived Cyclic Peptide Rt-D3
Organism: Chaetomium thermophilum, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-07-14 Classification: PROTEIN TRANSPORT/INHIBITOR |
 |
Crystal Structure Of Human Vps29 Complexed With Rapid-Derived Cyclic Peptide Rt-L2
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2021-07-14 Classification: PROTEIN TRANSPORT/INHIBITOR Ligands: SO4 |
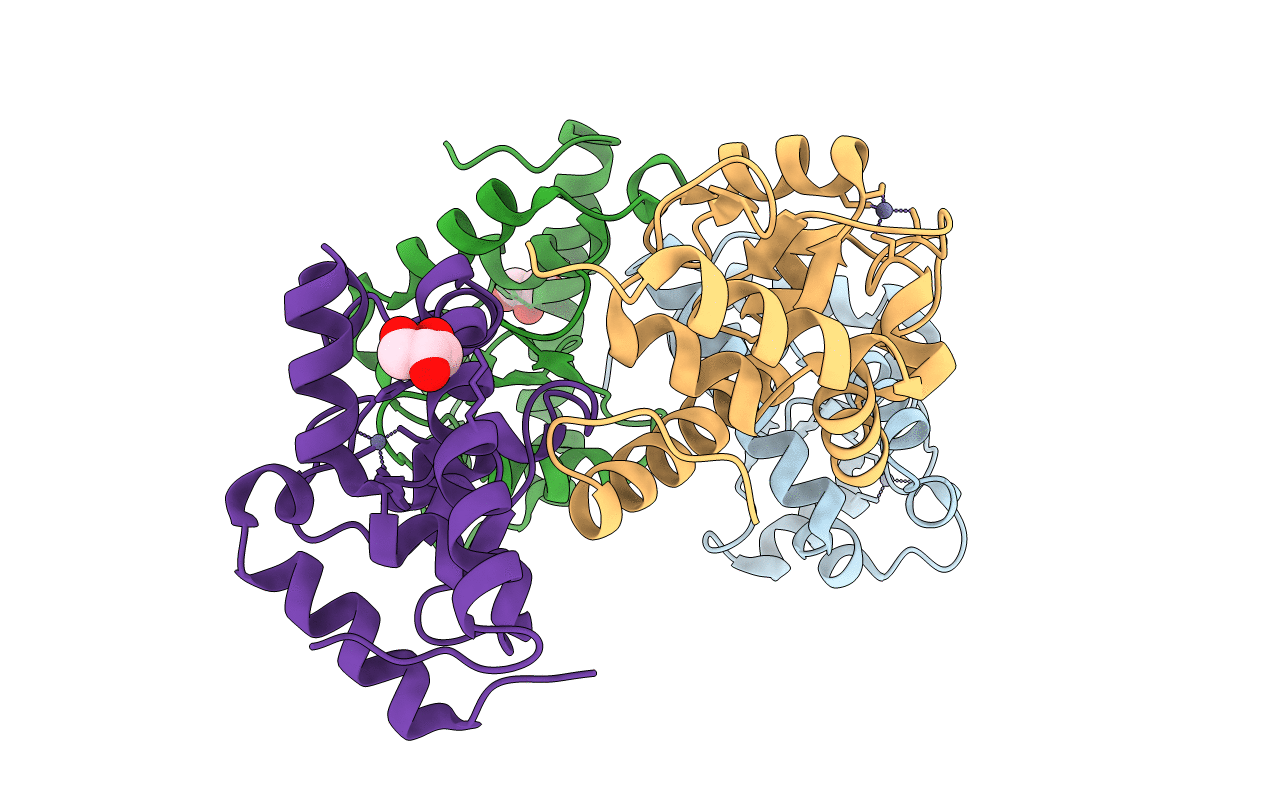 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2021-03-24 Classification: PROTEIN TRANSPORT Ligands: ZN, GOL |
 |
Organism: Human immunodeficiency virus type 1 group m subtype b (isolate bh10), Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-03-17 Classification: viral protein/RNA Ligands: MG, BOG |
 |
Structure Of Hiv-1 Reverse Transcriptase Initiation Complex Core With Efavirenz
Organism: Human immunodeficiency virus type 1 group m subtype b (isolate bh10), Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-03-17 Classification: viral protein/RNA Ligands: MG, EFZ |
 |
Structure Of Hiv-1 Reverse Transcriptase Initiation Complex Core With Nevirapine
Organism: Human immunodeficiency virus type 1 group m subtype b (isolate bh10), Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-03-17 Classification: viral protein/RNA Ligands: MG, NVP |
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:3.52 Å Release Date: 2020-12-16 Classification: CELL CYCLE |
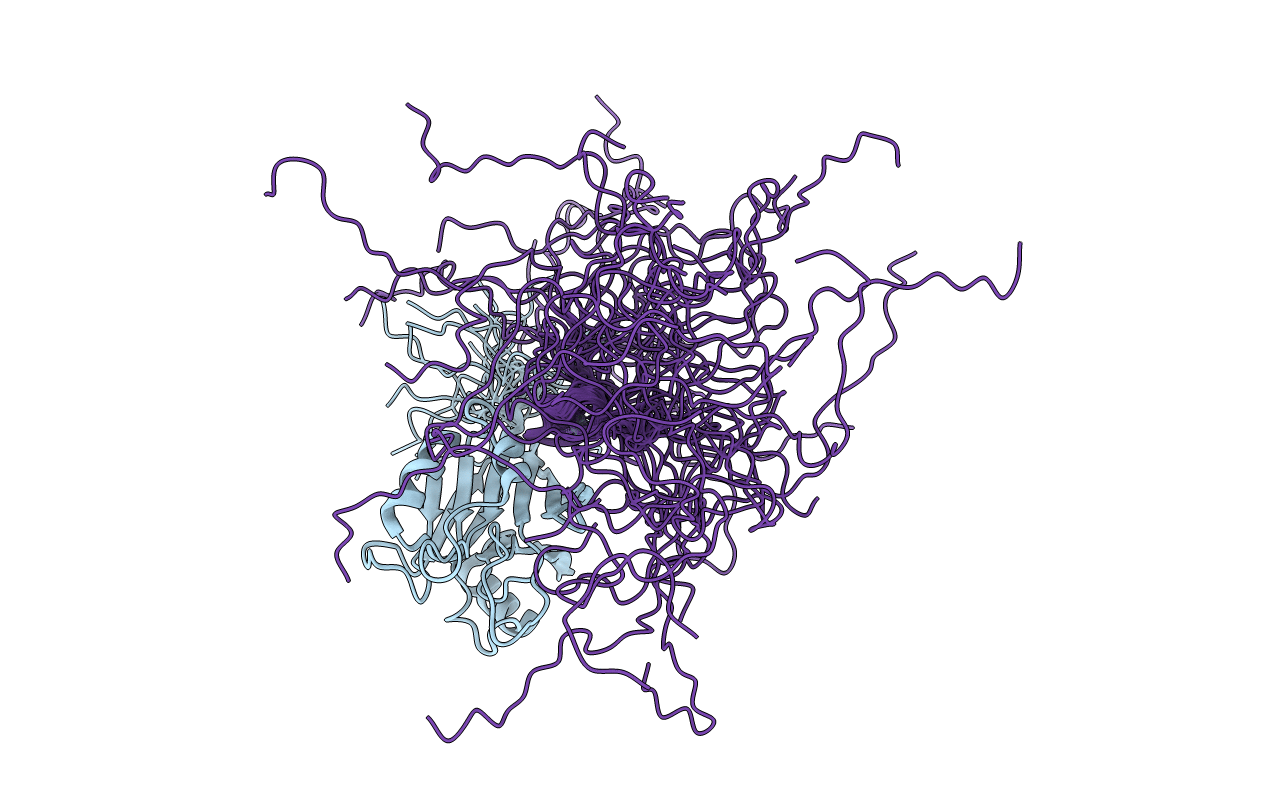 |
Solution Structure And 1H, 13C And 15N Chemical Shift Assignments For The Complex Of Vps29 With Varp 687-747
Organism: Mus musculus, Homo sapiens
Method: SOLUTION NMR Release Date: 2020-10-21 Classification: INTRACELLULAR VESICLE TRAFFICKING Ligands: ZN |
 |
+1 Extended Hiv-1 Reverse Transcriptase Initiation Complex Core (Pre-Translocation State)
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-06-24 Classification: VIRAL PROTEIN/RNA |
 |
+3 Extended Hiv-1 Reverse Transcriptase Initiation Complex Core (Pre-Translocation State)
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-06-24 Classification: VIRAL PROTEIN/RNA |
 |
+3 Extended Hiv-1 Reverse Transcriptase Initiation Complex Core (Intermediate State)
Organism: Human immunodeficiency virus type 1 group m subtype b (isolate bh10), Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-06-24 Classification: VIRAL PROTEIN/RNA |
 |
+3 Extended Hiv-1 Reverse Transcriptase Initiation Complex Core (Displaced State)
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-06-24 Classification: VIRAL PROTEIN/RNA |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2020-02-19 Classification: PROTEIN TRANSPORT |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2020-02-19 Classification: PROTEIN TRANSPORT |
 |
Organism: Geobacillus stearothermophilus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2019-06-05 Classification: TRANSFERASE/DNA Ligands: MPD, SO4, MG |
 |
Organism: Geobacillus stearothermophilus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2019-06-05 Classification: TRANSFERASE/DNA Ligands: SO4 |

