Search Count: 889
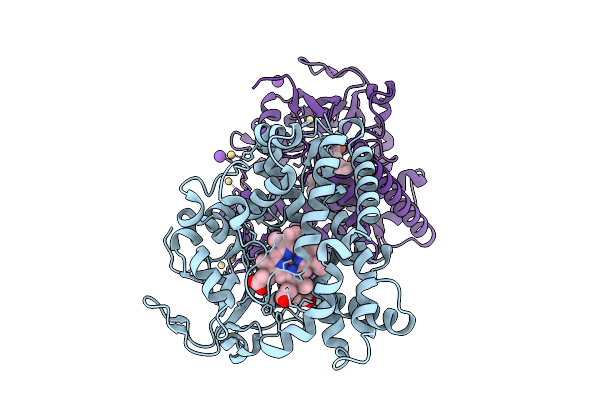 |
Crystal Structure Of The Cyp154C5 F92A/R114A/T248D/E282A Variant From Nocardia Farcinica
Organism: Nocardia farcinica ifm 10152
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: TES, HEM, CD, CL, K, GOL |
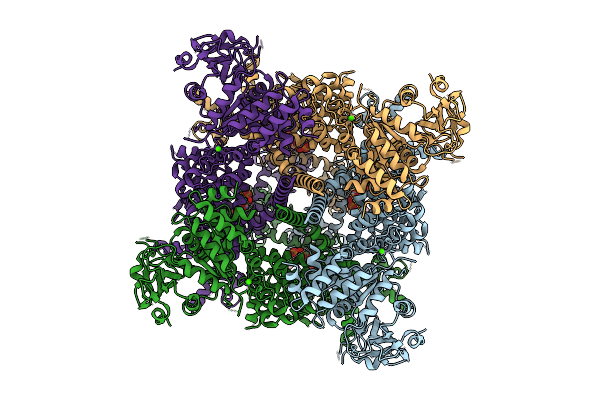 |
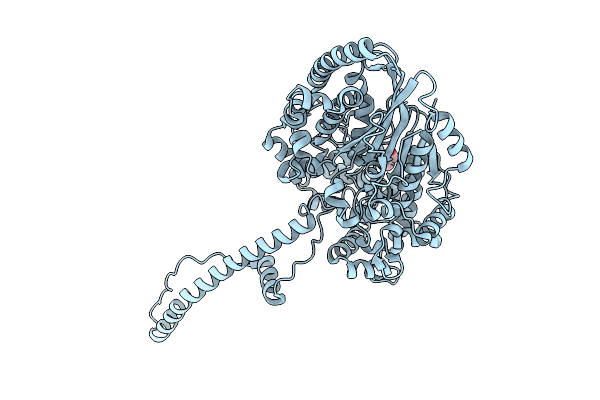
Cryo-Em Structure Of The Human Trpm4 Channel In A Calcium And Pi(4,5)P2 Bound Open State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: TRANSPORT PROTEIN Ligands: PT5, CA |
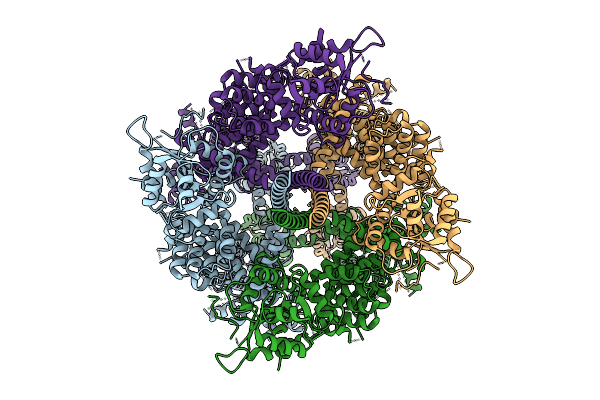 |
Cryo-Em Structure Of The Human Trpm4 Channel In A Calcium Bound Putative Desensitized State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: TRANSPORT PROTEIN Ligands: CA |
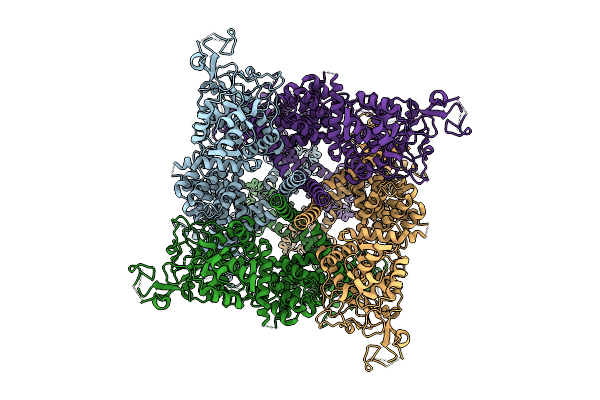 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: TRANSPORT PROTEIN |
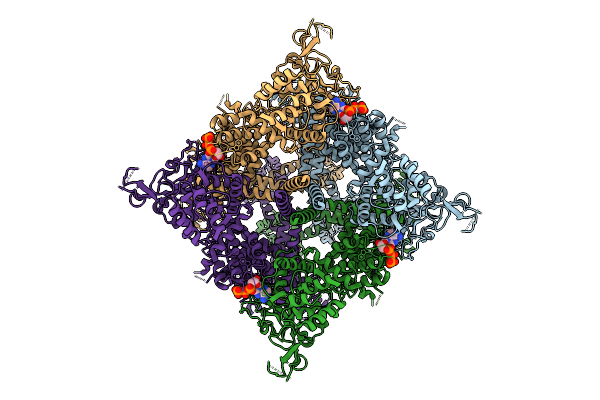 |
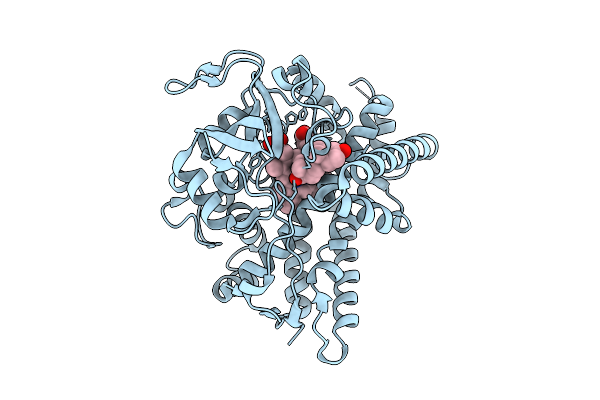
Cryo-Em Structure Of The Human Trpm4 Channel In A Atp Bound Inhibited State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: TRANSPORT PROTEIN Ligands: ATP |
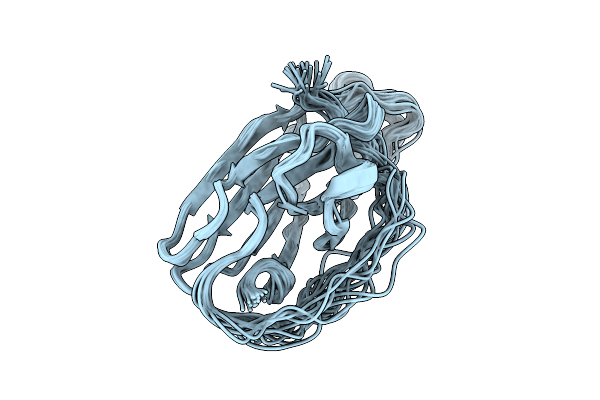 |
Organism: Thermus thermophilus
Method: SOLUTION NMR Release Date: 2025-10-22 Classification: STRUCTURAL PROTEIN |
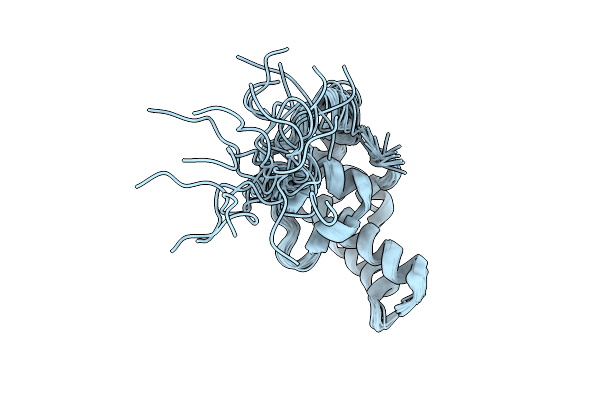 |
Organism: Thermus thermophilus
Method: SOLUTION NMR Release Date: 2025-10-22 Classification: STRUCTURAL PROTEIN |
 |
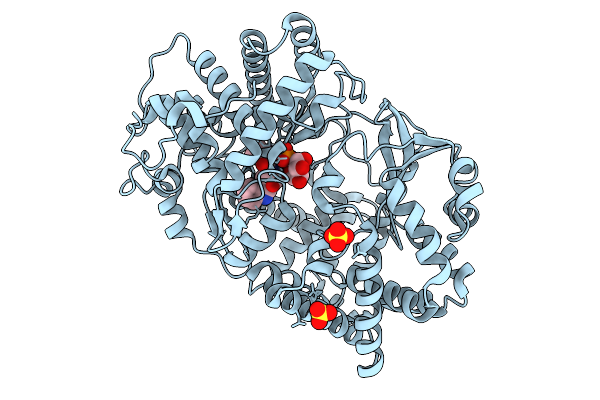
Crystal Structure Of The Cyp154C5 F92A/R114A/T248G/E282A Variant From Nocardia Farcinica
Organism: Nocardia farcinica ifm 10152
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: OXIDOREDUCTASE Ligands: HEM, TES |
 |
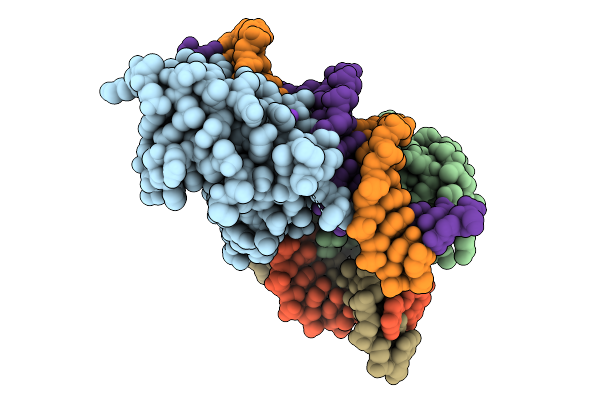
Crystallization Of Zinc Metalloproteinase Pepo From Porphyromonas Gingivalis
Organism: Porphyromonas gingivalis w83
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: METAL BINDING PROTEIN Ligands: RDF, ZN, SO4 |
 |
Co-Crystal Structure Of Yeast Forkhead Transcription Factor Fkh1 Bound To Dna
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: DNA BINDING PROTEIN/DNA Ligands: K |
 |
Cryo-Em Structure Of Histamine-Bound Histamine Receptor 3 H3R G Protein Complex
Organism: Homo sapiens, Bos taurus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: MEMBRANE PROTEIN Ligands: HSM |
 |
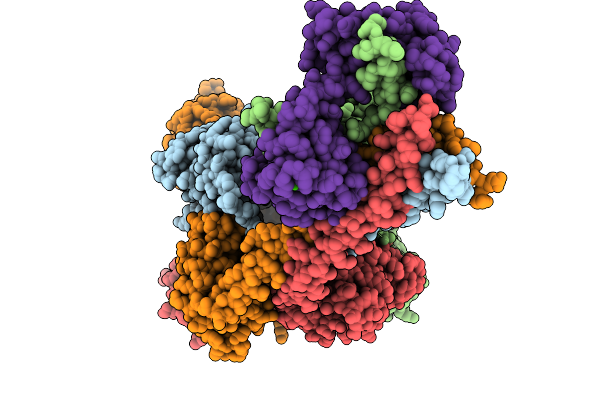
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: MEMBRANE PROTEIN Ligands: CA, CMP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
Cryo-Em Structure Of Histamine-Bound Histamine Receptor 4 H4R G Protein Complex
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: HSM, PO4 |
 |
Cryo-Em Structure Of Sars-Cov-2 Prototype Spike Protein In Complex With Triple-Nab 3G5, 4H5 And 4C11
Organism: Macaca mulatta, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 Prototype Spike Protein In Complex With Triple-Nab 4H1, 4A5 And 4C1
Organism: Macaca mulatta, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
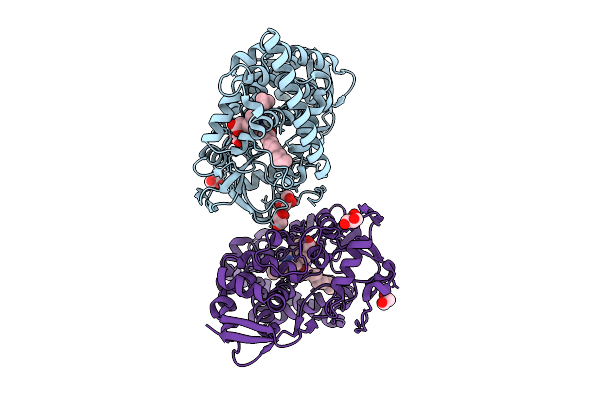
Cryo-Em Structure Of Sars-Cov-2 Eg.1 Spike Protein In Complex With Triple-Nab 4A5, 4C1 And 2E10
Organism: Macaca mulatta, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Crystal Structure Of The Cyp153A Double Mutant L354T/V456G From Marinobacter Aquaeolei
Organism: Marinobacter nauticus
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: HEM, DAO, GOL |
 |
Organism: Xanthomonas oryzae pv. oryzae pxo99a
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: TOXIN Ligands: GOL |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2025-08-06 Classification: DNA Ligands: ATP |

