Search Count: 56
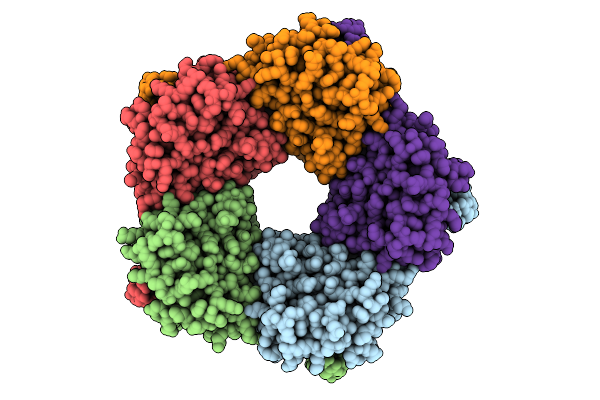 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE |
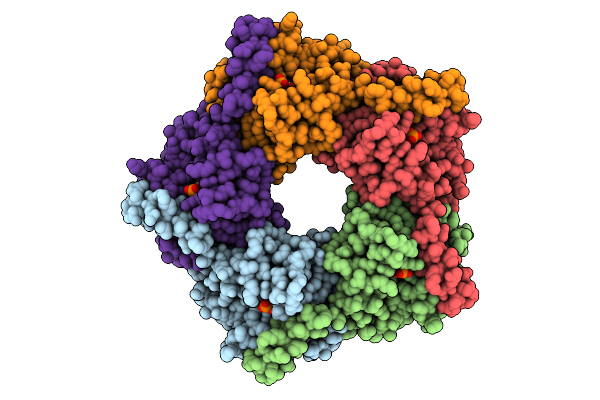 |
X-Ray Structure Of Enterobacter Cloaca Transaldolase In Complex With D-Fructose-6-Phosphate.
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: F6R |
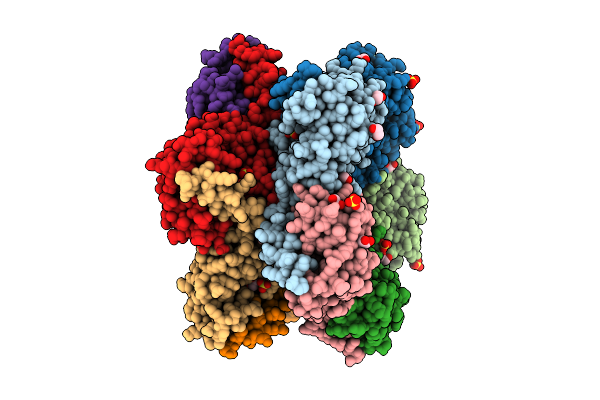 |
Organism: Thermus thermophilus (strain atcc 27634 / dsm 579 / hb8)
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: SO4, EDO |
 |
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-03-13 Classification: ISOMERASE Ligands: MN |
 |
Crystal Structure Of Lactobacillus Rhamnosus L-Rhamnose Isomerase In Complex With L-Rhamnose
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2024-03-13 Classification: ISOMERASE Ligands: MN, RM4, RAM |
 |
Crystal Structure Of Lactobacillus Rhamnosus L-Rhamnose Isomerase In Complex With D-Allulose
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2024-03-13 Classification: ISOMERASE Ligands: MN, PSJ, PSV |
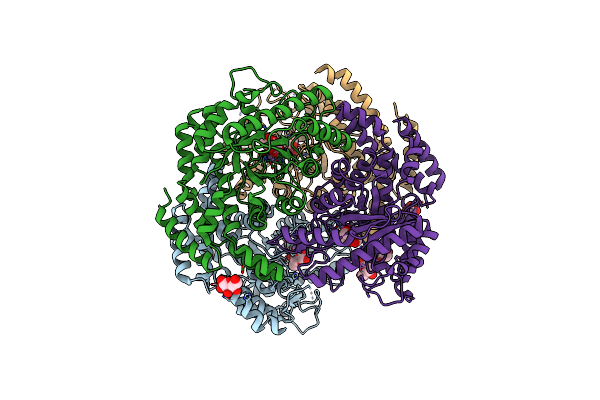 |
Crystal Structure Of Lactobacillus Rhamnosus L-Rhamnose Isomerase In Complex With D-Allose
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2024-03-13 Classification: ISOMERASE Ligands: MN, ALL, AFD |
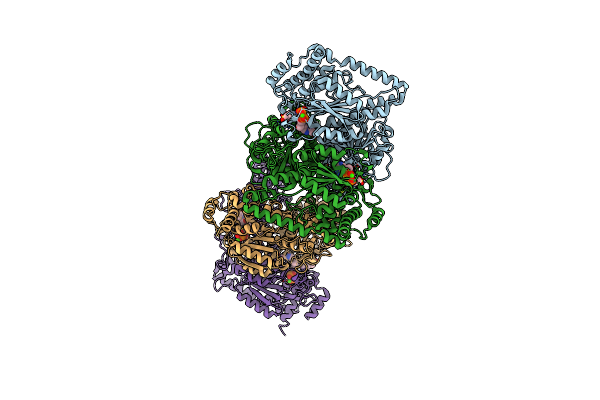 |
X-Ray Structure Of Thermus Thermophilus Hb8 Transketorase In Complex With Tpp And Mes
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2022-12-07 Classification: TRANSFERASE Ligands: TPP, CA, MES |
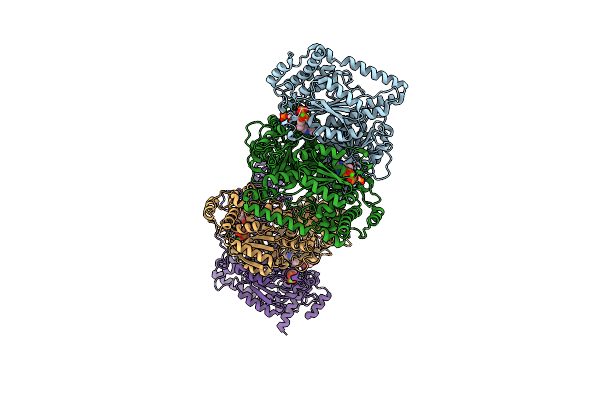 |
X-Ray Structure Ofthermus Thermophilus Hb8 Transketorase Demonstrate In Complex With Tpp And D-Erythrose-4-Phosphate
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-12-07 Classification: TRANSFERASE Ligands: TPP, CA, E4P |
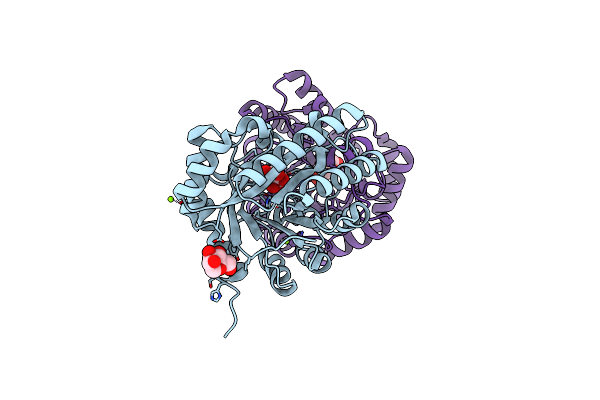 |
Crystal Structure Of Homo Dimeric D-Allulose 3-Epimerase From Methylomonas Sp. In Complex With D-Fructose
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-04-28 Classification: ISOMERASE Ligands: MN, FUD, MG, BDF |
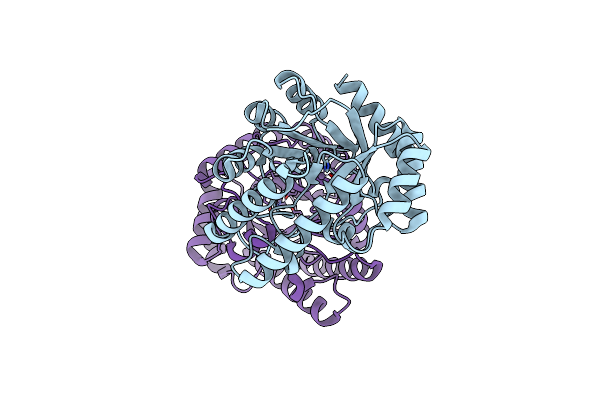 |
Crystal Structure Of Homo Dimeric D-Allulose 3-Epimerase From Methylomonas Sp.
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2021-04-21 Classification: ISOMERASE Ligands: MN |
 |
Crystal Structure Of Homo Dimeric D-Allulose 3-Epimerase From Methylomonas Sp. In Complex With D-Allulose
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-04-21 Classification: ISOMERASE Ligands: MN, PSJ |
 |
Crystal Structure Of Homo Dimeric D-Allulose 3-Epimerase From Methylomonas Sp. In Complex With L-Tagatose
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-04-21 Classification: ISOMERASE Ligands: SOL, MN, MG, FZU, LTG |
 |
Crystal Structure Of N-Terminal His-Tagged D-Allulose 3-Epimerase From Methylomonas Sp. In Complex With D-Allulose
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2021-04-21 Classification: ISOMERASE Ligands: MN, EPE, PSJ |
 |
Crystal Structure Of N-Terminal His-Tagged D-Allulose 3-Epimerase From Methylomonas Sp. With Additional C-Terminal Residues
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2021-04-21 Classification: ISOMERASE Ligands: MN, FUD, EPE, EDO |
 |
Crystal Structure Of Arthrobacter Globiformis M30 Sugar Epimerase Which Can Produce D-Allulose From D-Fructose
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2018-10-24 Classification: ISOMERASE Ligands: MN, ACT |
 |
Crystal Structure Of D-Tagatose 3-Epimerase C66S From Pseudomonas Cichorii In Complex With 1-Deoxy L-Tagatose, Using A Crystal Grown In Microgravity
Organism: Pseudomonas cichorii
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2016-04-27 Classification: ISOMERASE Ligands: MN, TGJ, TGK |
 |
Crystal Structure Of D-Tagatose 3-Epimerase C66S From Pseudomonas Cichorii In Complex With 1-Deoxy D-Tagatose
Organism: Pseudomonas cichorii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-03-23 Classification: ISOMERASE Ligands: MN, TGD, TGY |
 |
Crystal Structure Of D-Tagatose 3-Epimerase C66S From Pseudomonas Cichorii In Complex With 1-Deoxy L-Tagatose
Organism: Pseudomonas cichorii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-03-23 Classification: ISOMERASE Ligands: MN, TGK, TGJ |
 |
Crystal Structure Of D-Tagatose 3-Epimerase C66S From Pseudomonas Cichorii In Complex With 1-Deoxy 3-Keto D-Galactitol
Organism: Pseudomonas cichorii
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2016-03-23 Classification: ISOMERASE Ligands: MN, TGS, TGR |

