Search Count: 225
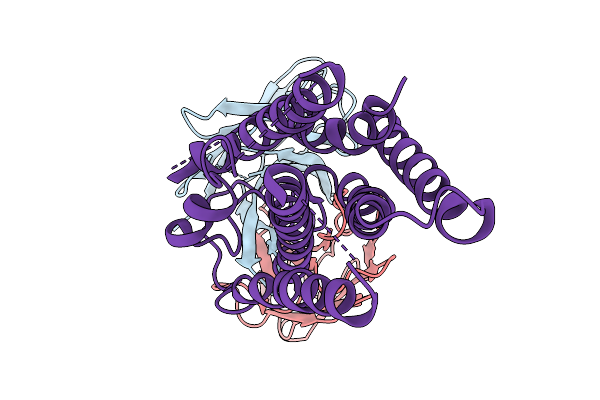 |
Cryo-Em Structure Of The Human A2A Adenosine Receptor In Complex With A Fab Antibody Fragment
Organism: Mus musculus, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.45 Å Release Date: 2026-01-07 Classification: MEMBRANE PROTEIN |
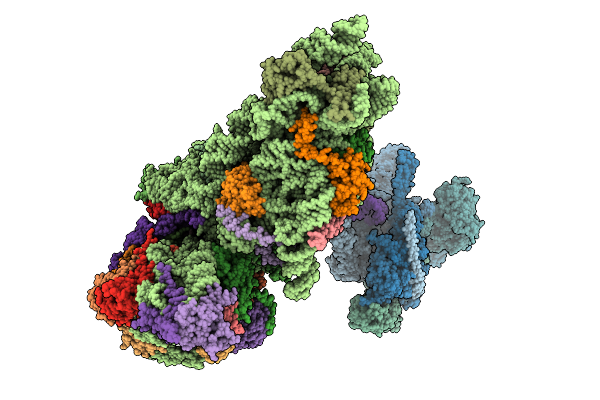 |
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG |
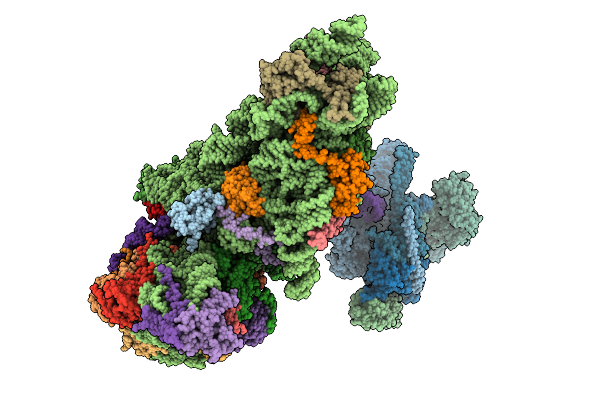 |
Structure Of The Human 40S Ribosome Complexed With Hcv Ires, Eif1A And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG |
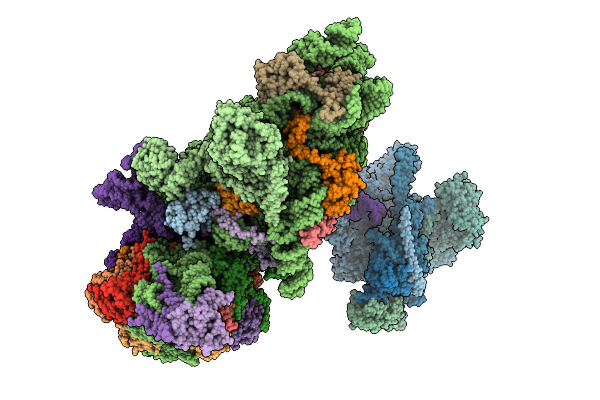 |
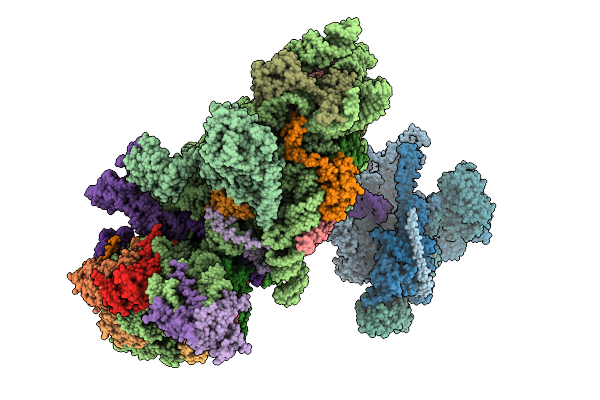
Structure Of The Hcv Ires-Dependent Pre-48S Translation Initiation Complex With Eif1A, Eif5B, And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, GTP, ZN |
 |
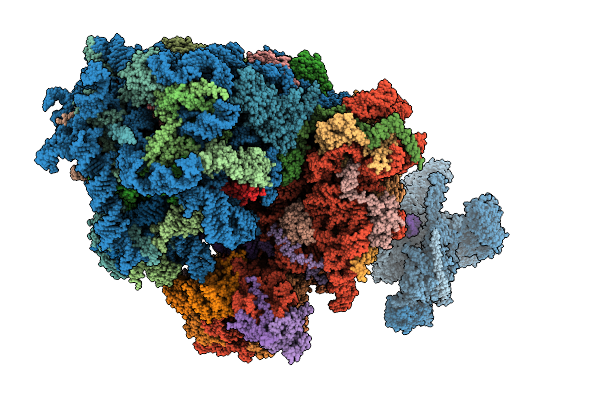
Structure Of The Hcv Ires-Dependent 48S Translation Initiation Complex With Eif5B And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG, GTP |
 |
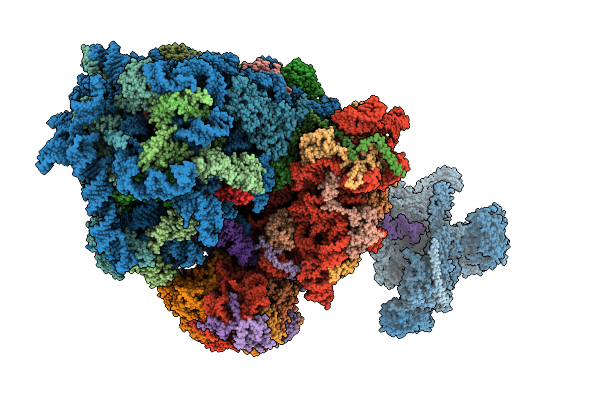
Cryo-Em Structure Of The Hcv Ires-Dependently Initiated Cmv-Stalled 80S Ribosome (Non-Rotated State) In Complexed With Eif3
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, ZN |
 |
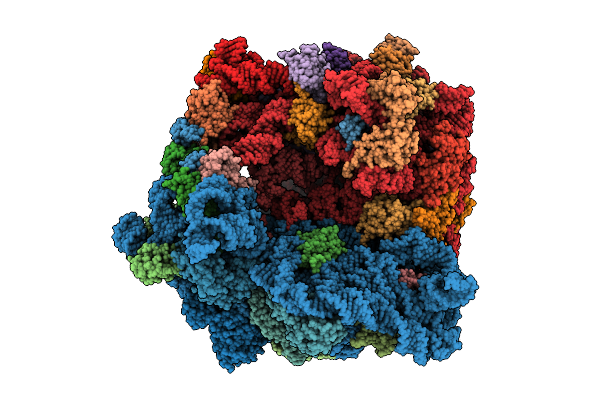
Cryo-Em Structure Of The Hcv Ires-Dependently Initiated Cmv-Stalled 80S Ribosome (Rotated State) In Complexed With Eif3
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, ZN |
 |
Structure Of The Bacterial Ribosome Without Hypoxia-Induced Rrna Modifications
Organism: Escherichia phage t4, Escherichia coli bw25113
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RIBOSOME Ligands: MG |
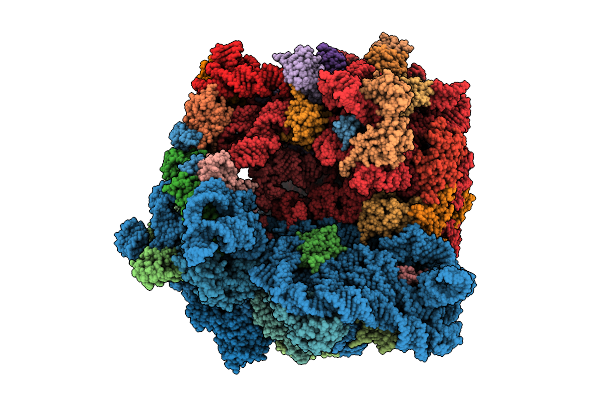 |
Structure Of The Bacterial Ribosome With Hypoxia-Induced Rrna Modifications
Organism: Escherichia phage t4, Escherichia coli bw25113
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RIBOSOME Ligands: MG |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2025-10-29 Classification: HYDROLASE Ligands: GLU, ACT, GOL |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-10-29 Classification: HYDROLASE Ligands: ACT, GOL |
 |
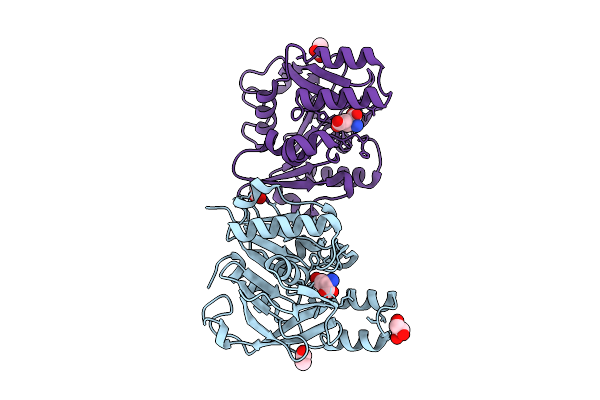
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2025-10-29 Classification: HYDROLASE Ligands: ACT, GOL |
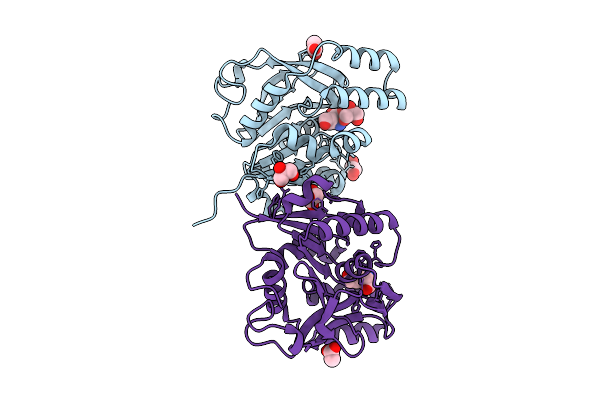 |
Gamma-Glutamyl Peptidase 1 From Arabidopsis Thaliana (H192N Gamma-Glu Intermediate)
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2025-10-29 Classification: HYDROLASE Ligands: GLU, ACT, GOL |
 |
Cryo-Em Structure Of Tmprss2 In Complex With Fab Fragments Of 752 Mab And 2228 Mab
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN |
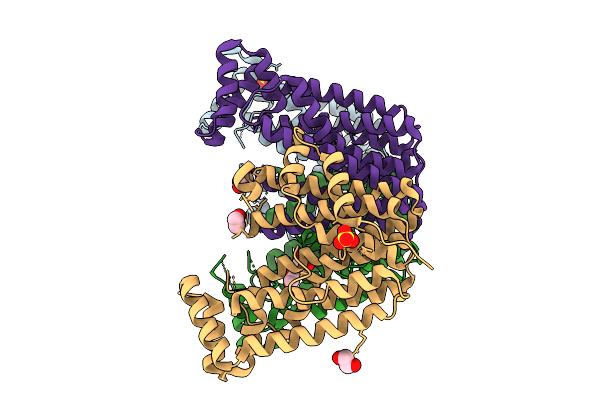 |
Organism: Streptomyces sp. ks84
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: SO4, EDO |
 |
Organism: Streptomyces sp. ks84
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: FPS, MG |
 |
Organism: Streptomyces sp. ks84
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: MG |
 |
Organism: Streptomyces sp. ks84
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: SO4, EDO, FPS, MG |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:3.12 Å Release Date: 2025-07-16 Classification: PLANT PROTEIN |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2025-07-16 Classification: PLANT PROTEIN |

