Search Count: 80,965
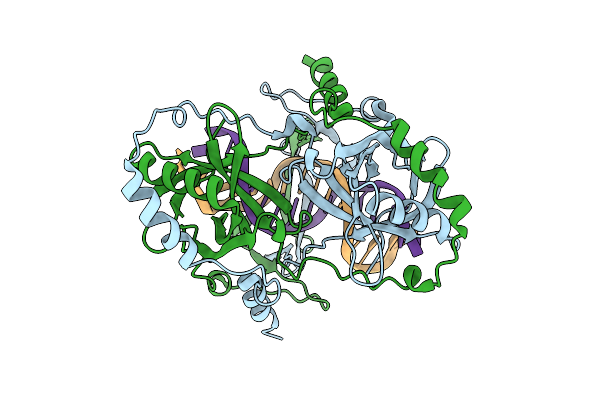 |
Organism: Mycobacterium tuberculosis h37rv, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN |
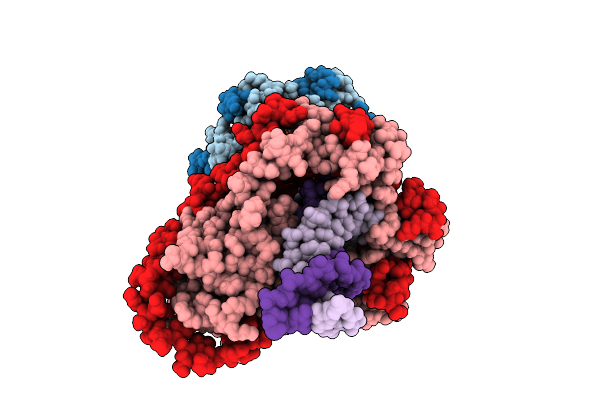 |
Organism: Mycobacterium tuberculosis h37rv, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN/DNA |
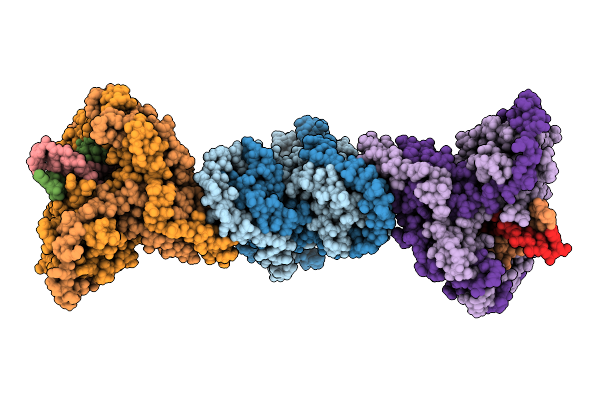 |
Organism: Mycobacterium tuberculosis h37rv, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN |
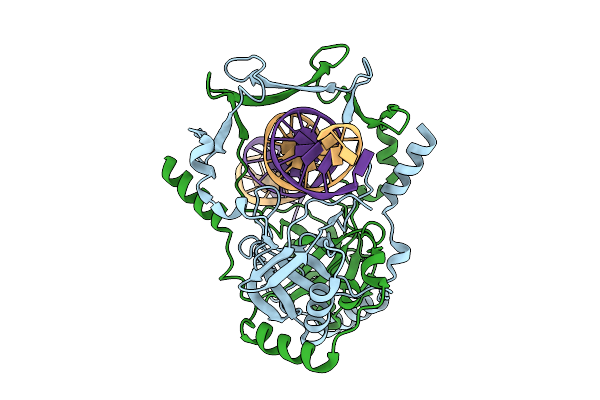 |
Organism: Mycobacterium tuberculosis h37rv, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN |
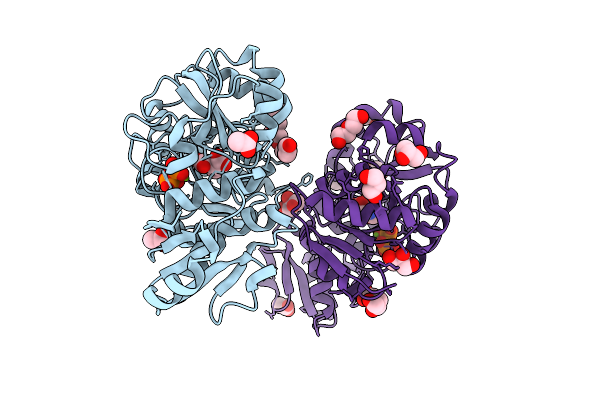 |
Thermotoga Maritima Threonylcarbamoyl Adenylate Synthase (Tsac2) In Complex With N-Carboxy-L-Threonine, Magnesium And Pyrophosphate.
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: U6A, MG, DPO, P6G, ACT, GOL, PEG |
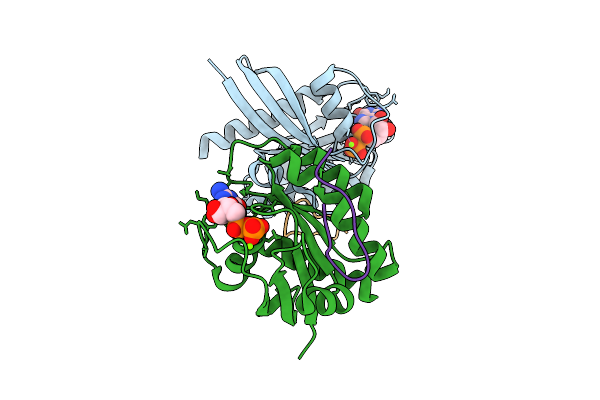 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MG, GDP |
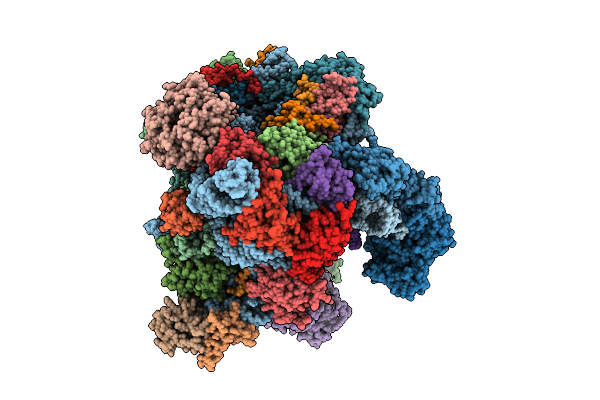 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSLATION Ligands: SF4 |
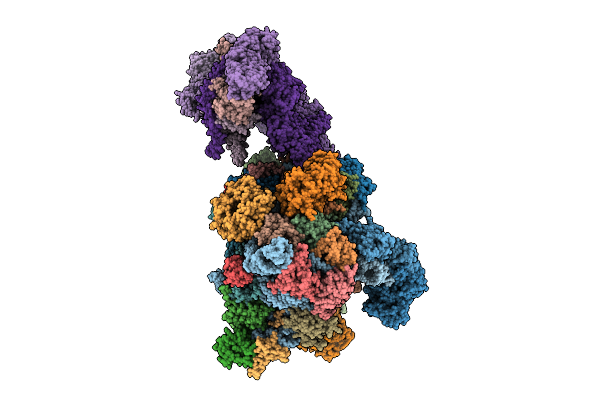 |
Late-Stage 48S Initiation Complex With Eif3 (Ls48S-Eif3 Ic) Guided By The Trans-Rna
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSLATION Ligands: SF4 |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2025-12-10 Classification: DNA Ligands: LDP |
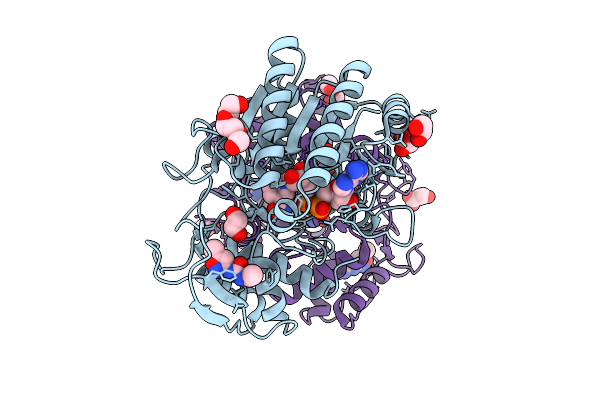 |
Structure Of Udp-Galactose-4-Epimerase (Gale) Bound To Fragment From Diamond Xchem Experiment.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: CARBOHYDRATE Ligands: NAD, JGA, MLI, PGE, EDO, CL |
 |
Solution Structure Of An Intramolecular Anti-Parallel G-Quadruplex With A 5'-End Overhang.
|
 |
Organism: Mycobacterium tuberculosis, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN |
 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: GOL, PEG |
 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: A1L6P, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: ISOMERASE Ligands: A1EHL, PE8, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: ISOMERASE Ligands: A1EFQ, PE8, SO4 |
 |
Organism: Thermus phage philo
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: PEPTIDE NUCLEIC ACID |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ANTIVIRAL PROTEIN/DNA/RNA Ligands: ATP, ZN, MG |

