Search Count: 15
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2023-11-22 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2023-11-22 Classification: DE NOVO PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-10-13 Classification: OXYGEN TRANSPORT Ligands: CMO, HEM |
 |
Crystal Structure Of Equus Ferus Caballus Glutathione Transferase A3-3 In Complex With Glutathione
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-11-18 Classification: TRANSFERASE Ligands: GSH, EDO |
 |
Crystal Structure Of Equus Ferus Caballus Glutathione Transferase A3-3 In Complex With Glutathione And Triethyltin
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-11-18 Classification: TRANSFERASE Ligands: GSH, QLT, MPD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-03-13 Classification: TRANSCRIPTION Ligands: HV4 |
 |
Crystal Structure Of A V2-Directed, Rv144 Vaccine-Like Antibody From Hiv-1 Infection, Cap228-16H, Bound To A Heterologous V2 Peptide
Organism: Homo sapiens, Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2018-11-21 Classification: IMMUNE SYSTEM Ligands: SO4 |
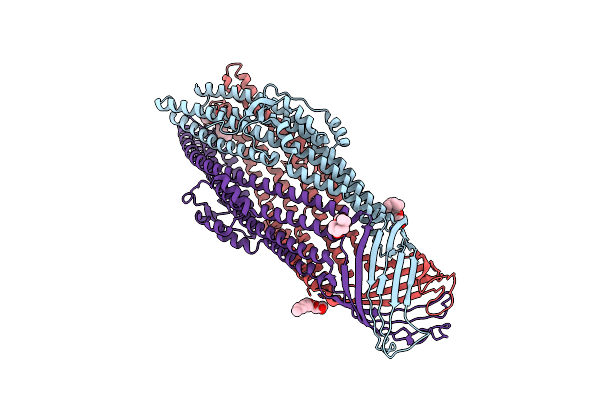 |
Crystal Structure Of An Antigenic Outer Membrane Protein St50 From Salmonella Typhi
Organism: Salmonella enterica subsp. enterica serovar typhi
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2015-12-23 Classification: MEMBRANE PROTEIN Ligands: BOG |
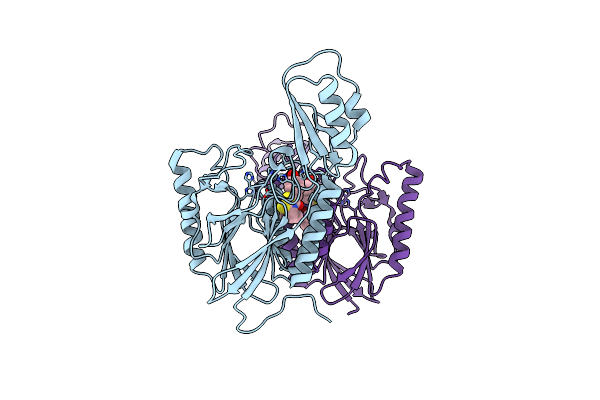 |
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-01-15 Classification: DNA BINDING PROTEIN/inhibitor Ligands: 2Q0, MN |
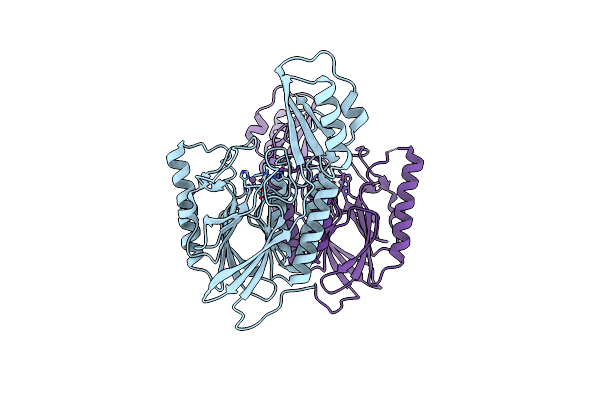 |
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-01-08 Classification: HYDROLASE Ligands: MN |
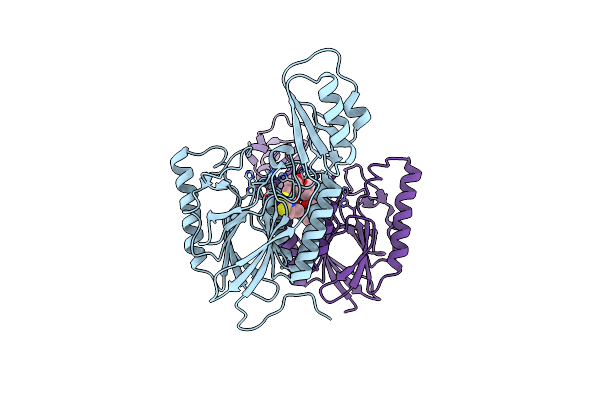 |
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-01-08 Classification: DNA BINDING PROTEIN/inhibitor Ligands: MN, 2PW |
 |
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-01-08 Classification: DNA BINDING PROTEIN/inhibitor Ligands: MN, 2PK |
 |
Dna Double-Strand Break Repair Pathway Choice Is Directed By Distinct Mre11 Nuclease Activities
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-01-08 Classification: DNA BINDING PROTEIN/inhibitor Ligands: MN, 2PV |
 |
Structure Of The C-Terminal Truncated Form Of E.Coli C5-Hydroxylase Ubii Involved In Ubiquinone (Q8) Biosynthesis
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-05-29 Classification: OXIDOREDUCTASE Ligands: GOL, CL, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2010-12-01 Classification: PROTEIN BINDING Ligands: TRS, CL, GOL |

