Search Count: 35
 |
Neutron Structure Of Gh1 Beta-Glucosidase Td2F2 Ligand-Free Form At Room Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: NHE, DOD |
 |
Neutron Structure Of Gh1 Beta-Glucosidase Td2F2 Glucose Complex At Room Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: BGC, NHE, DOD |
 |
Neutron Structure Of Gh1 Beta-Glucosidase Td2F2 2F-Glc Complex At Room Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: G2F, NHE, DOD |
 |
X-Ray Structure Of Gh1 Beta-Glucosidase Td2F2 2F-Glc Complex At Room Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: G2F, NHE |
 |
X-Ray Structure Of Gh1 Beta-Glucosidase Td2F2 2F-Glc Complex At Cryogenic Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: G2F, NA, GOL, NHE |
 |
Crystal Structure Of Group 1 Oligosaccharide-Releasing Beta-N-Acetylgalactosaminidase Ngaca From Cohnella Abietis, Apo 1 Form
Organism: Cohnella abietis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: MG |
 |
Crystal Structure Of Group 1 Oligosaccharide-Releasing Beta-N-Acetylgalactosaminidase Ngaca From Cohnella Abietis, Apo 2 Form
Organism: Cohnella abietis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: MG |
 |
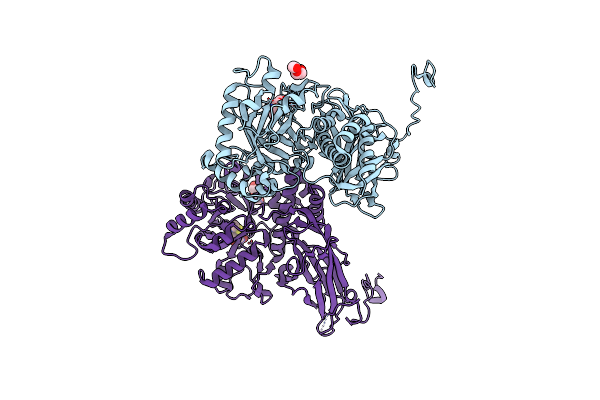
Crystal Structure Of Group 2Oligosaccharide/Monosaccharide-Releasing Beta-N-Acetylhexosaminidase Ngaat From Arabidopsis Thaliana In Complex With Galnac-Thiazoline
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: GNL, PEG, CL |
 |
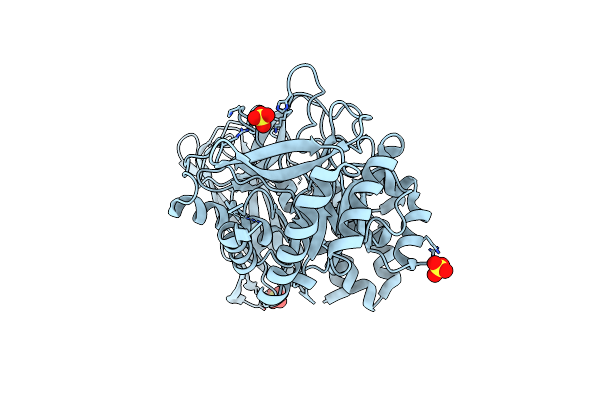
Crystal Structure Of Group 2 Oligosaccharide/Monosaccharide-Releasing Beta-N-Acetylhexosaminidase Ngaat From Arabidopsis Thaliana In Complex With Glcnac-Thiazoline
Organism: Arabidopsis thaliana x arabidopsis lyrata
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: PEG, NGT, CL |
 |
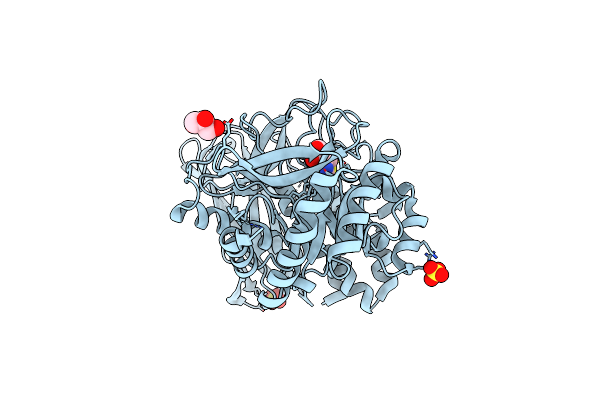
Crystal Structure Of Group 3 Oligosaccharide/Monosaccharide-Releasing Beta-N-Acetylgalactosaminidase Ngadssm, Apo Form
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: SO4 |
 |
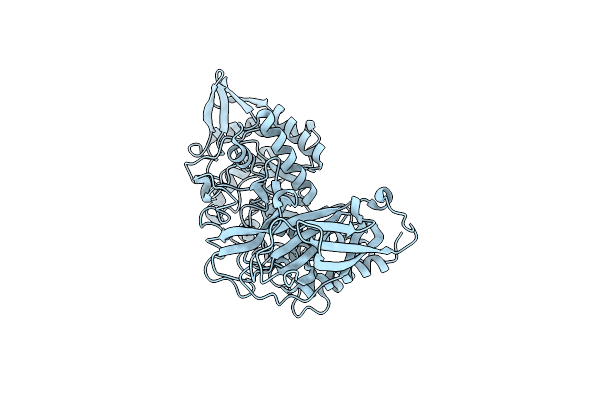
Crystal Structure Of Group 3 Oligosaccharide/Monosaccharide-Releasing Beta-N-Acetylgalactosaminidase Ngadssm In Complex With Galnac-Thiazoline
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: GNL, GOL, SO4, CL |
 |
Crystal Structure Of Group 4 Monosaccharide-Releasing Beta-N-Acetylgalactosaminidase Ngap2 From Paenibacillus Sp. Ts12, Apo Form
Organism: Paenibacillus sp. ts12
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: BR |
 |
Crystal Structure Of Group 4 Monosaccharide-Releasing Beta-N-Acetylgalactosaminidase Ngap2 From Paenibacillus Sp. Ts12 In Complex With Galnac-Thiazoline
Organism: Paenibacillus sp. ts12
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-04-24 Classification: HYDROLASE Ligands: GNL, BR |
 |
Crystal Structure Of Group 4 Monosaccharide-Releasing Beta-N-Acetylgalactosaminidase Ngaly From Lacticaseibacillus Yichunensis, Apo Form
Organism: Lacticaseibacillus yichunensis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-04-24 Classification: HYDROLASE |
 |
Crystal Structure Of Gh146 Beta-L-Arabinofuranosidase Bll3Hypba1 (Amino Acids 380-1223) In Complex With Tris
Organism: Bifidobacterium longum subsp. longum jcm 1217
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-02-21 Classification: HYDROLASE Ligands: ZN, TRS, NA, GOL, MG |
 |
Crystal Structure Of Gh146 Beta-L-Arabinofuranosidase Bll3Hypba1 (Amino Acids 380-1051), Ligand-Free Form
Organism: Bifidobacterium longum subsp. longum jcm 1217
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-02-21 Classification: HYDROLASE Ligands: ZN |
 |
Beta-L-Arabinofurano-Cyclitol Aziridines Are Cysteine-Directed Broad-Spectrum Inhibitors And Activity-Based Probes For Retaining Beta-L-Arabinofuranosidases
Organism: Bifidobacterium longum
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-12-13 Classification: HYDROLASE Ligands: ZN, UI5 |
 |
Gh146 Beta-L-Arabinofuranosidase From Bacteroides Thetaioatomicron In Complex With Beta-L-Arabinofurano Cyclophellitol Aziridine
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-12-13 Classification: HYDROLASE Ligands: UI5, ZN, UHU |
 |
Endo-Alpha-D-Arabinanase Endoma1 From Microbacterium Arabinogalactanolyticum
Organism: Microbacterium arabinogalactanolyticum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-08-16 Classification: HYDROLASE |
 |
Endo-Alpha-D-Arabinanase Endoma1 D51N Mutant From Microbacterium Arabinogalactanolyticum In Complex With Arabinooligosaccharides
Organism: Microbacterium arabinogalactanolyticum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: GOL, PGE, PEG, NA, CA, OYO, MES |

