Search Count: 48
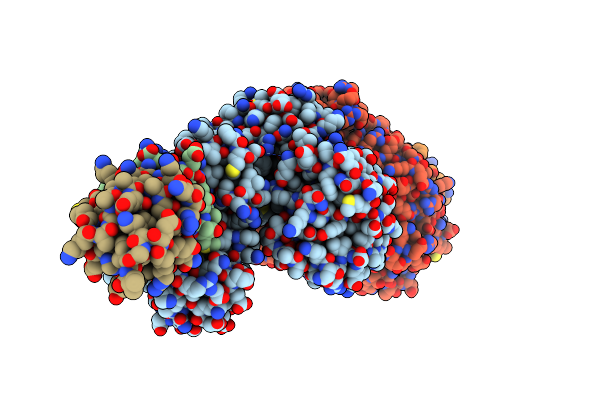 |
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-10-06 Classification: DNA BINDING PROTEIN/REPLICATION |
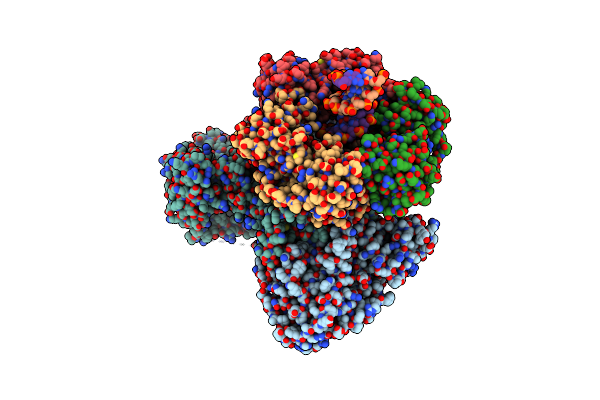 |
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1), Thermococcus kodakarensis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-08-05 Classification: REPLICATION/DNA Ligands: FE, ZN |
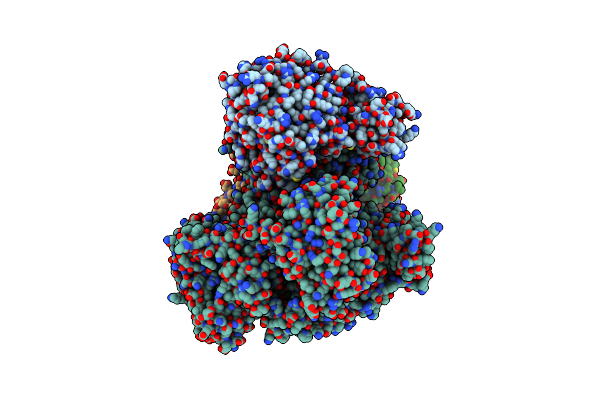 |
Organism: Thermococcus kodakarensis kod1, Thermococcus kodakarensis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-08-05 Classification: REPLICATION/DNA Ligands: FE, ZN |
 |
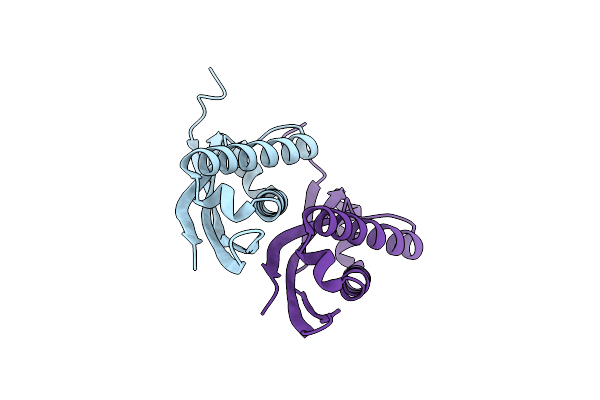
Crystal Structure Of Archaeal Ribosomal Protein Ap1, Apelota, And Gtp-Bound Aef1A Complex
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-11-13 Classification: TRANSLATION Ligands: GTP, MG, NA |
 |
Organism: Vibrio alginolyticus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-03-06 Classification: MOTOR PROTEIN |
 |
Organism: Vibrio alginolyticus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2019-03-06 Classification: MOTOR PROTEIN |
 |
Crystal Structure Of The Novel Lesion-Specific Endonuclease Pfuendoq From Pyrococcus Furiosus
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-04-04 Classification: DNA BINDING PROTEIN Ligands: ZN, SM |
 |
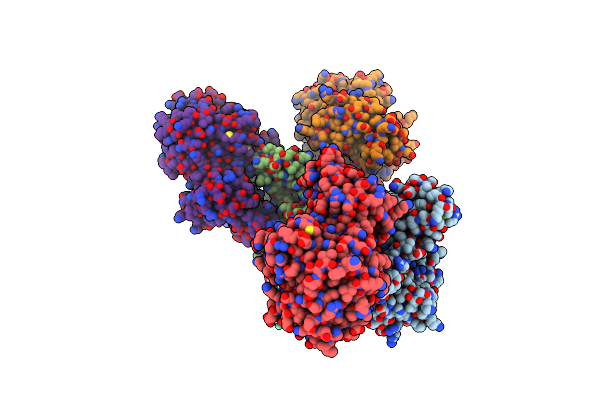
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MG, MPD |
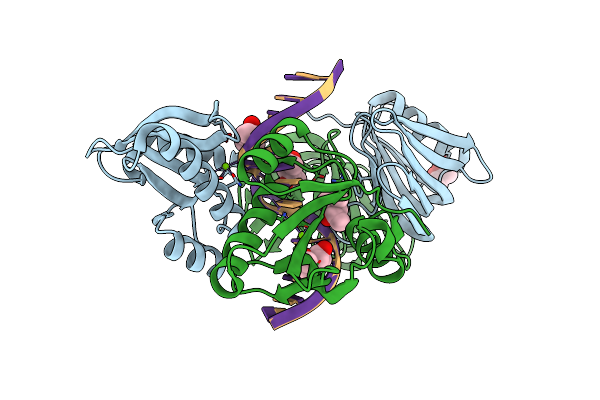 |
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MPD, MG |
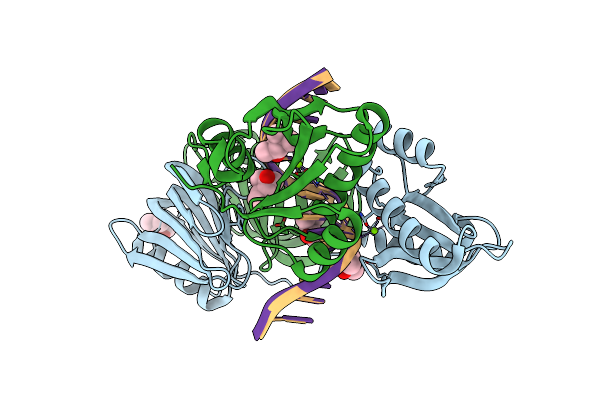 |
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MG, MPD |
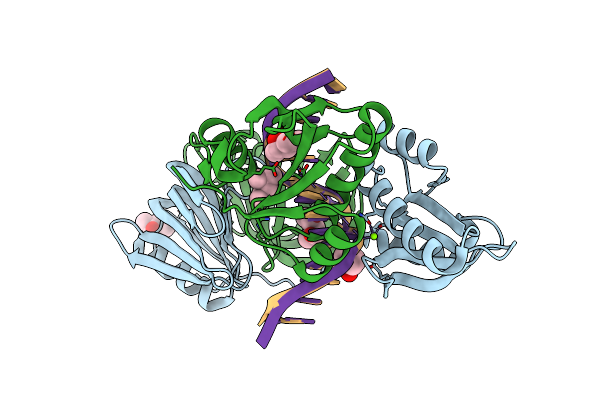 |
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MPD, MG |
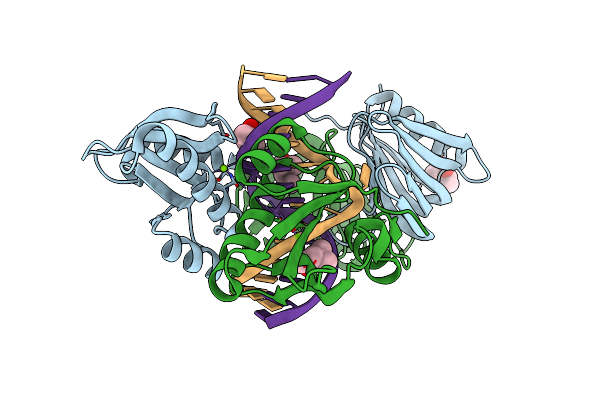 |
Organism: Thermococcus kodakarensis kod1, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-11-02 Classification: HYDROLASE/DNA Ligands: MG, MPD |
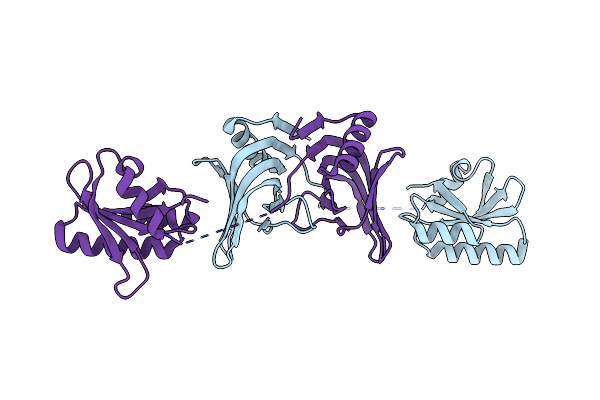 |
Organism: Thermococcus kodakarensis kod1
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-11-02 Classification: HYDROLASE |
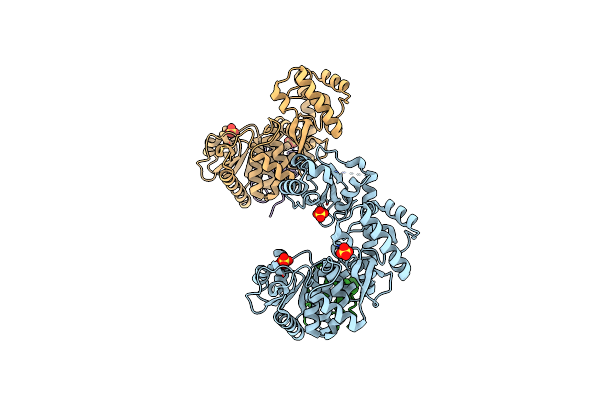 |
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2016-10-12 Classification: DNA BINDING PROTEIN/REPLICATION Ligands: SO4 |
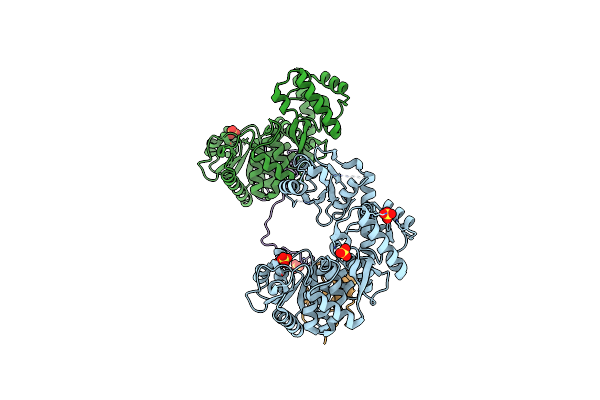 |
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1), Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2016-10-12 Classification: DNA BINDING PROTEIN/REPLICATION Ligands: SO4 |
 |
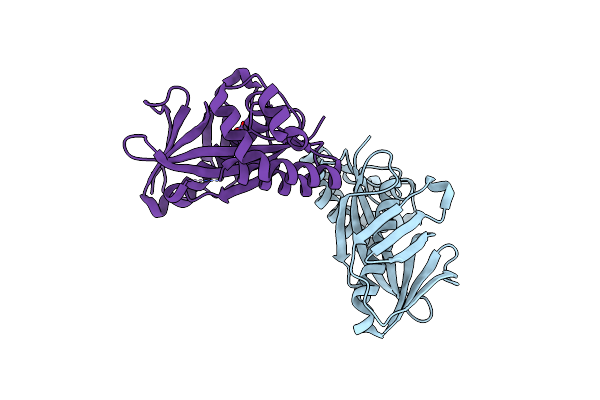
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-10-12 Classification: DNA BINDING PROTEIN |
 |
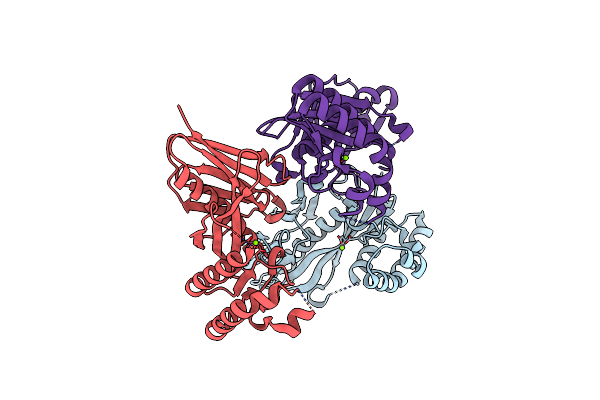
Crystal Structure Of A Trimeric Exonuclease Phoexo I From Pyrococcus Horikoshii Ot3 At 1.52A Resolution.
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2015-07-15 Classification: HYDROLASE Ligands: MG |
 |
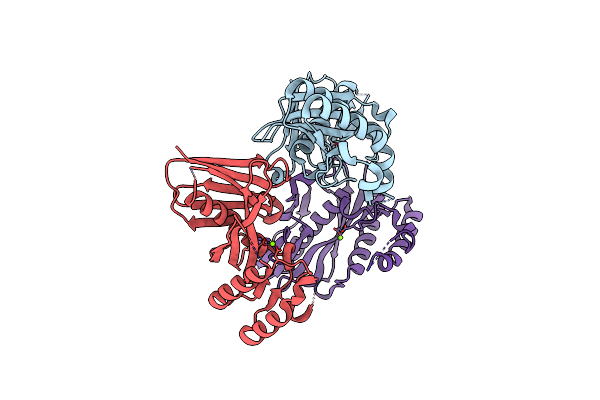
Crystal Structure Of A Trimeric Exonuclease Phoexo I From Pyrococcus Horikoshii Ot3 At 2.15A Resolution
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2015-07-15 Classification: HYDROLASE Ligands: MG |
 |
Crystal Structure Of A Trimeric Exonuclease Phoexo I From Pyrococcus Horikoshii Ot3 At 2.20A Resolution.
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-07-15 Classification: HYDROLASE Ligands: MG |
 |
Crystal Structure Of A Trimeric Exonuclease Phoexo I From Pyrococcus Horikoshii Ot3 In Complex With Poly-Da
Organism: Pyrococcus horikoshii, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2015-07-15 Classification: HYDROLASE/DNA |

