Search Count: 21
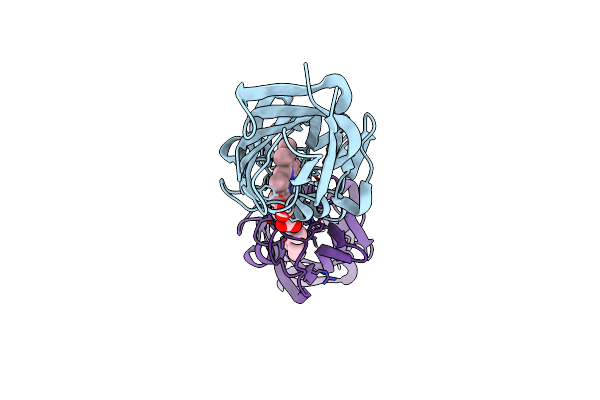 |
Crystal Structure Of The N-Terminal Domain Of Htaa From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / jcm 1318 / lmg 3730 / ncimb 10025)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-11-20 Classification: HEME-BINDING PROTEIN Ligands: HEM |
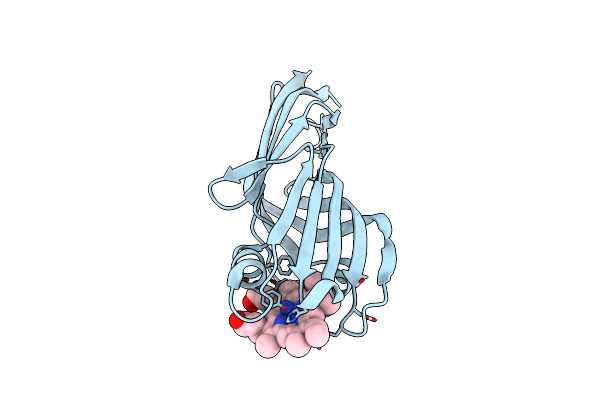 |
Crystal Structure Of The C-Terminal Domain Of Htaa From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / jcm 1318 / lmg 3730 / ncimb 10025)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-11-20 Classification: HEME-BINDING PROTEIN Ligands: HEM |
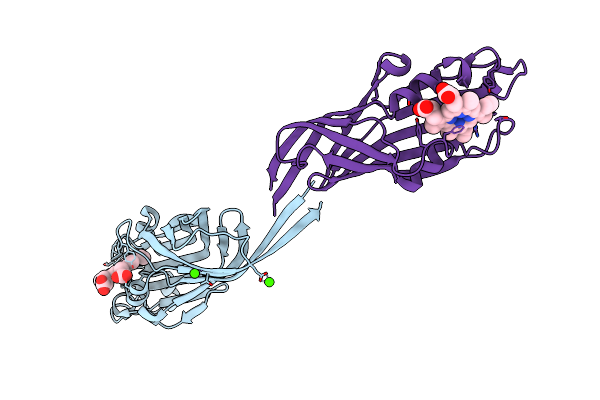 |
Crystal Structure Of Heme Binding Protein Htab From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / jcm 1318 / lmg 3730 / ncimb 10025)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-11-20 Classification: HEME-BINDING PROTEIN Ligands: HEM, CA |
 |
Crystal Structure Of The H434A Variant Of The C-Terminal Domain Of Htaa From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / jcm 1318 / lmg 3730 / ncimb 10025)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-11-20 Classification: HEME-BINDING PROTEIN Ligands: HEM, NA |
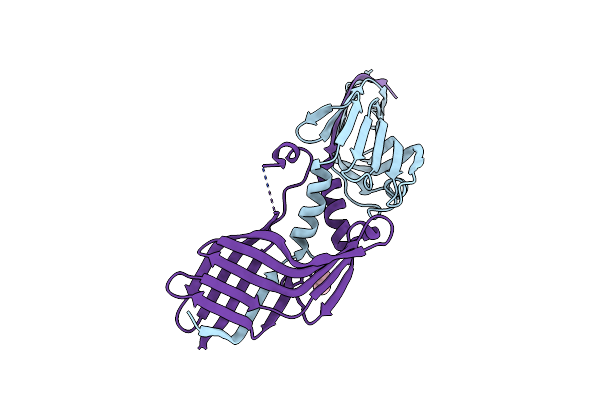 |
Crystal Structure Of The Domain-Swapped Dimer H434A Variant Of The C-Terminal Domain Of Htaa From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / jcm 1318 / lmg 3730 / ncimb 10025)
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2019-11-20 Classification: HEME-BINDING PROTEIN Ligands: IPA |
 |
Solution Structure Of Peptidyl-Prolyl Cis/Trans Isomerase Domain Of Trigger Factor In Complex With Mbp
Organism: Escherichia coli k-12
Method: SOLUTION NMR Release Date: 2018-08-22 Classification: CHAPERONE |
 |
Organism: Escherichia coli (strain k12)
Method: SOLUTION NMR Release Date: 2018-06-13 Classification: CHAPERONE |
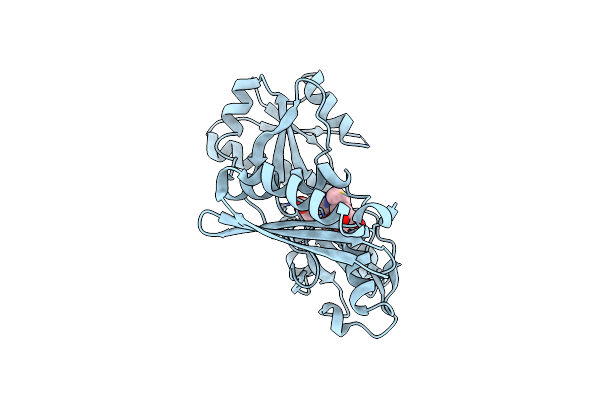 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-11-15 Classification: TRANSFERASE Ligands: DPM, MG |
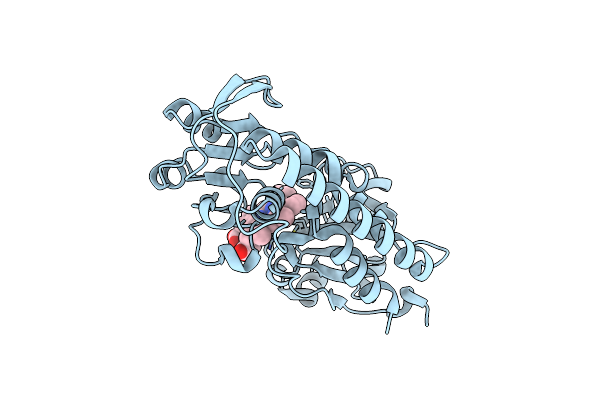 |
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2017-03-01 Classification: TRANSPORT PROTEIN Ligands: HEM |
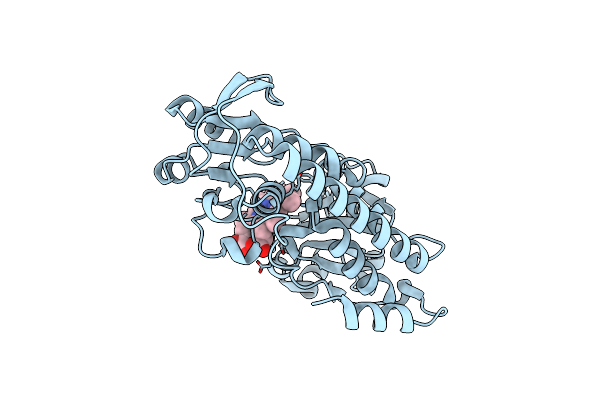 |
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2017-03-01 Classification: TRANSPORT PROTEIN Ligands: HEM |
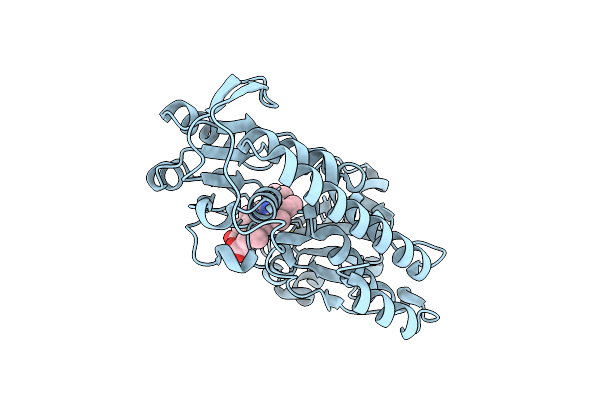 |
Organism: Corynebacterium glutamicum atcc 13032
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2017-03-01 Classification: TRANSPORT PROTEIN Ligands: HEM |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2016-07-13 Classification: HEME-BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-03-23 Classification: MEMBRANE PROTEIN Ligands: HEM |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2016-02-17 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2016-02-17 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2015-10-21 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Crystal Structure Of The Ferrous Co-Bound Cytochrome P450Cam Mutant (L358P/C334A)
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-06-01 Classification: OXIDOREDUCTASE Ligands: K, HEM, CMO, CAM |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-05-25 Classification: OXIDOREDUCTASE Ligands: K, HEM, CAM |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-05-25 Classification: OXIDOREDUCTASE Ligands: K, HEM, CAM, CMO, TRS |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-05-25 Classification: OXIDOREDUCTASE Ligands: K, HEM, CAM, TRS |

