Search Count: 40
 |
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2024-12-04 Classification: MOTOR PROTEIN |
 |
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: MOTOR PROTEIN |
 |
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: MOTOR PROTEIN |
 |
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: MOTOR PROTEIN |
 |
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-06-19 Classification: TRANSFERASE |
 |
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-06-19 Classification: TRANSFERASE Ligands: MG, GTP |
 |
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-06-19 Classification: TRANSFERASE |
 |
Crystal Structure Of Bovine Heart Cytochrome C Oxidase, Apo Structure With Dmso
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-12-21 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PER, PGV, TGL, CUA, CDL, CHD, PEK, PSC, ZN, DMU |
 |
Crystal Structure Of Bovine Heart Cytochrome C Oxidase, The Structure Complexed With An Allosteric Inhibitor T113
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-12-21 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PER, TGL, J6X, CUA, CHD, PSC, PEK, PGV, CDL, DMU, ZN |
 |
Cryo-Em Structure Of Cytochrome Bo3 From Escherichia Coli, Apo Structure With Dmso
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-12-21 Classification: OXIDOREDUCTASE Ligands: HEO, HEM, CU, PEE, UNX |
 |
Cryo-Em Structure Of Cytochrome Bo3 From Escherichia Coli, The Structure Complexed With An Allosteric Inhibitor N4
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-12-21 Classification: OXIDOREDUCTASE Ligands: HEO, HEM, CU, PEE, JYR, UNX |
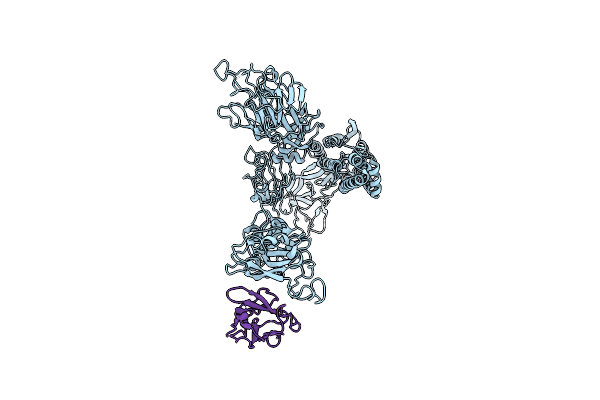 |
Minor Cryo-Em Structure Of S Protein Trimer Of Sars-Cov2 With K-874A Vhh, Composite Map
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: PROTEIN BINDING |
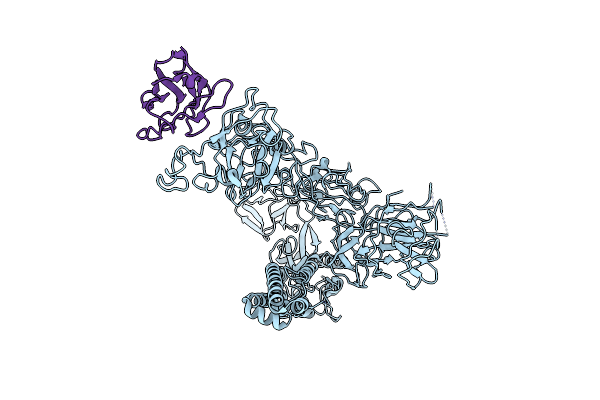 |
Major Cryo-Em Structure Of S Protein Trimer Of Sars-Cov2 With K-874, Composite Map
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: PROTEIN BINDING |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: PROTEIN BINDING |
 |
Organism: Synechococcus elongatus
Method: SOLUTION NMR Release Date: 2021-07-14 Classification: UNKNOWN FUNCTION |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2019-06-12 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2019-06-12 Classification: DE NOVO PROTEIN Ligands: SO4 |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2019-06-12 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2019-06-12 Classification: DE NOVO PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-05-08 Classification: HYDROLASE Ligands: ONC |

