Search Count: 12
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2004-10-05 Classification: HYDROLASE Ligands: FE, MG, GOL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2004-10-05 Classification: HYDROLASE Ligands: FE, MG, GOL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2004-10-05 Classification: HYDROLASE Ligands: FE, MG, GOL |
 |
Bacterial Cytosine Deaminase D314G Mutant Bound To 5-Fluoro-4-(S)-Hydroxy-3,4-Dihydropyrimidine.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2004-10-05 Classification: HYDROLASE Ligands: FE, FPY |
 |
Bacterial Cytosine Deaminase D314A Mutant Bound To 5-Fluoro-4-(S)-Hydroxyl-3,4-Dihydropyrimidine.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2004-10-05 Classification: HYDROLASE Ligands: FE, FPY, GOL |
 |
Bacterial Cytosine Deaminase D314S Mutant Bound To 5-Fluoro-4-(S)-Hydroxyl-3,4-Dihydropyrimidine.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2004-10-05 Classification: HYDROLASE Ligands: FE, FPY, GOL |
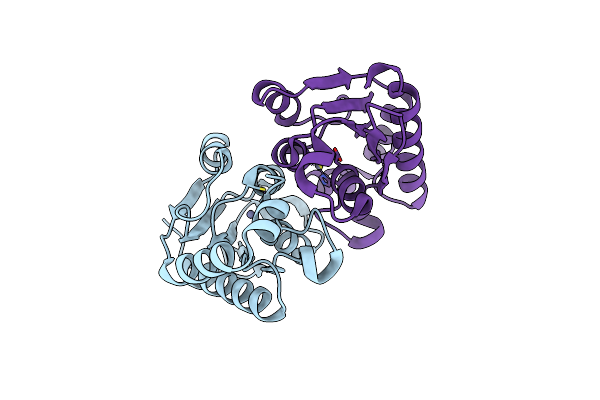 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-03-02 Classification: HYDROLASE Ligands: ZN |
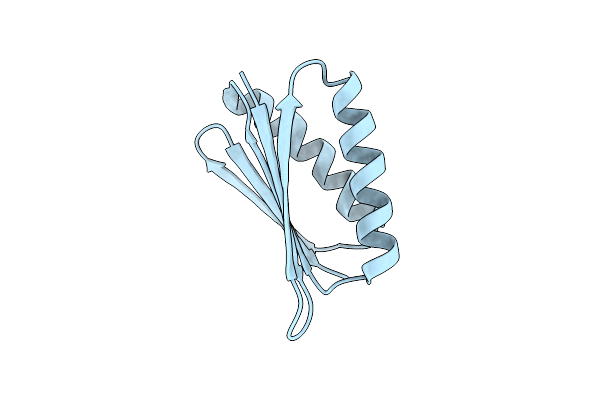 |
Crystal Structure Of Top7: A Computationally Designed Protein With A Novel Fold
Organism: Computationally designed sequence
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2003-11-25 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Yeast Cytosine Deaminase Apo-Enzyme: Inorganic Zinc Bound
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2003-08-19 Classification: HYDROLASE Ligands: ZN, CA |
 |
The Crystal Structure Of Yeast Cytosine Deaminase Bound To 4(R)-Hydroxyl-3,4-Dihydropyrimidine At 1.14 Angstroms.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2003-08-19 Classification: HYDROLASE Ligands: ZN, HPY, ACY, CA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2002-02-06 Classification: HYDROLASE Ligands: FE |
 |
The Structure Of Escherichia Coli Cytosine Deaminase Bound To 4-Hydroxy-3,4-Dihydro-1H-Pyrimidin-2-One
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2002-02-06 Classification: HYDROLASE Ligands: FE, HPY |

