Search Count: 19
 |
Organism: Kitasatospora cystarginea
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-06-26 Classification: BIOSYNTHETIC PROTEIN Ligands: PG4, FLC |
 |
Perdeuterated Tyrosine Phenol-Lyase From Citrobacter Freundii Complexed With An Aminoacrylate Intermediate Formed From S-Ethyl-L-Cysteine And 4-Hydroxypyridine
Organism: Citrobacter freundii
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2020-02-12 Classification: lyase/lyase inhibitor Ligands: K, CQG, 0JO, P33, DHA |
 |
Crystal Structure Of Nitrite Bound Synthetic Core Domain Of Nitrite Reductase From Ralstonia Pickettii (Residues 1-331)
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, PG4, NO2 |
 |
Crystal Structure Of As Isolated Synthetic Core Domain Of Nitrite Reductase From Ralstonia Pickettii (Residues 1-331)
Organism: Ralstonia sp. 5_2_56faa
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, CL, PG4 |
 |
Crystal Structure Of As Isolated Y323A Mutant Of Haem-Cu Containing Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, HEC |
 |
Crystal Structure Of Nitrite Bound Y323A Mutant Of Haem-Cu Containing Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, HEC, NO2 |
 |
Crystal Structure Of As Isolated Y323E Mutant Of Haem-Cu Containing Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, HEC, GOL |
 |
Crystal Structure Of Nitrite Bound Y323E Mutant Of Haem-Cu Containing Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, HEC, NO2, GOL |
 |
Crystal Structure Of As Isolated Y323F Mutant Of Haem-Cu Containing Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, HEC, GOL |
 |
Crystal Structure Of Nitrite Bound Y323F Mutant Of Haem-Cu Containing Nitrite Reductase From Ralstonia Pickettii
Organism: Ralstonia pickettii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-11-06 Classification: METAL BINDING PROTEIN Ligands: CU, NO2, HEC |
 |
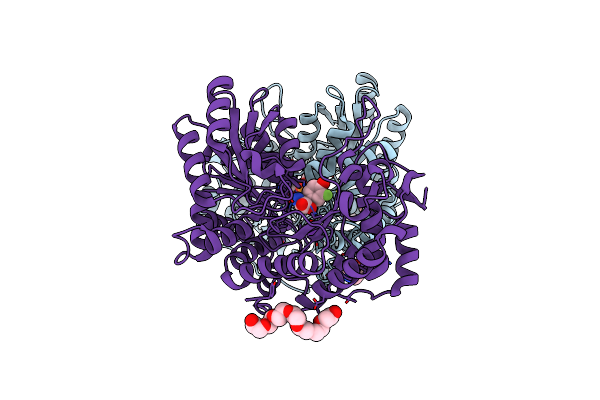
Citrobacter Freundii Tyrosine Phenol-Lyase Complexed With 4-Hydroxypyridine And Aminoacrylate From L-Serine
Organism: Citrobacter freundii
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2019-10-16 Classification: LYASE Ligands: CQG, 0JO, K, SER, P33 |
 |
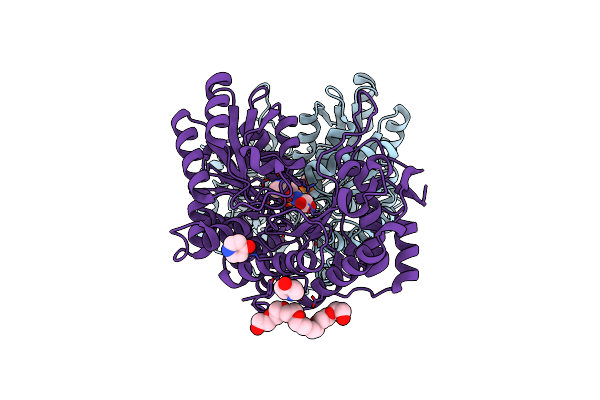
Citrobacter Freundii Tyrosine Phenol-Lyase Complexed With 4-Hydroxypyridine And Aminoacrylate From 3-F-L-Tyrosine
Organism: Citrobacter freundii
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2019-10-16 Classification: LYASE Ligands: K, CQG, 0JO, P33, YOF |
 |
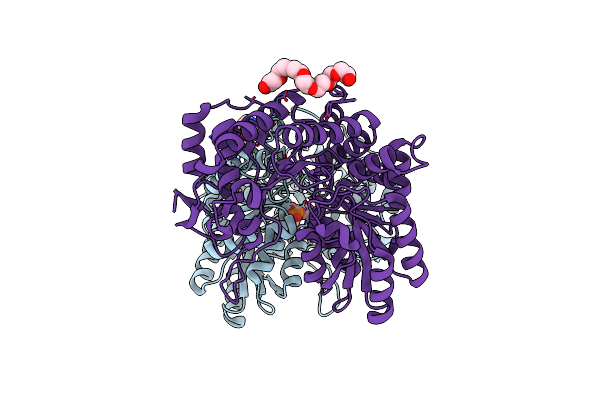
Citrobacter Freundii F448A Mutant Tyrosine Phenol-Lyase Complexed With 4-Hydroxypyridine And Aminoacrylate From S-Ethyl-L-Cysteine
Organism: Citrobacter freundii
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-10-16 Classification: LYASE Ligands: CQG, K, 0JO, P33 |
 |
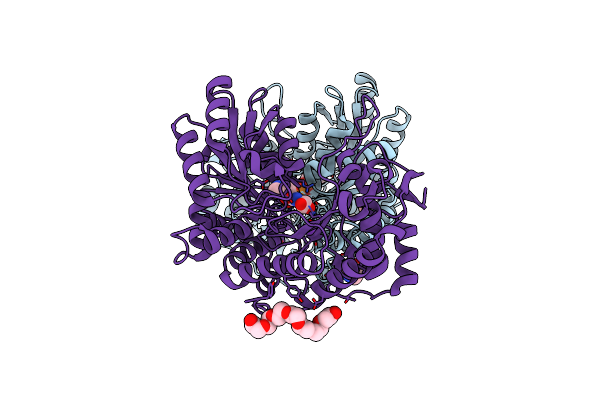
Citrobacter Freundii Tyrosine Phenol-Lyase Complexed With 4-Hydroxypyridine And Aminoacrylate From L-Tyrosine
Organism: Citrobacter freundii
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2019-10-02 Classification: LYASE Ligands: K, CQG, 0JO, P33 |
 |
Citrobacter Freundii Tyrosine Phenol-Lyase Complexed With 4-Hydroxypyridine And Aminoacrylate From S-Ethyl-L-Cysteine
Organism: Citrobacter freundii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-10-02 Classification: LYASE Ligands: K, CQG, 0JO, P33 |
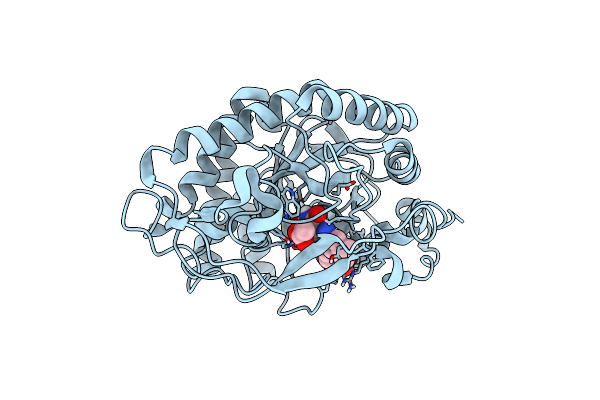 |
Crystal Structure Of Pentaerythritol Tetranitrate Reductase (Petnr) Mutant L25I
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-03-20 Classification: FLAVOPROTEIN Ligands: FMN, ACT |
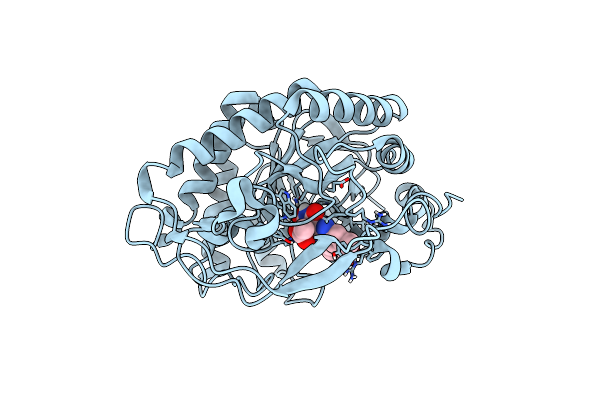 |
Crystal Structure Of Pentaerythritol Tetranitrate Reductase (Petnr) Mutant L25A
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2019-03-20 Classification: FLAVOPROTEIN Ligands: FMN, ACT |
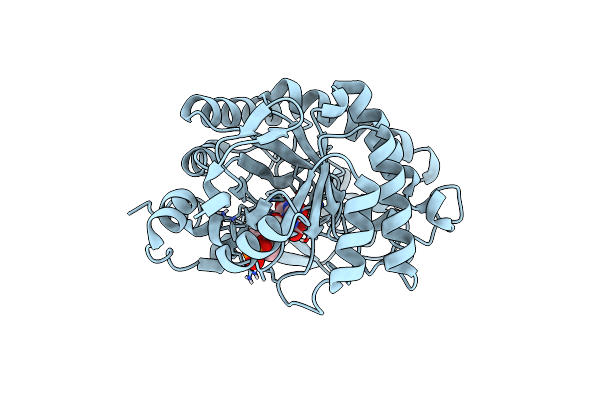 |
Crystal Structure Of Pentaerythritol Tetranitrate Reductase (Petnr) Mutant I107L
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-03-20 Classification: FLAVOPROTEIN Ligands: FMN, ACT |
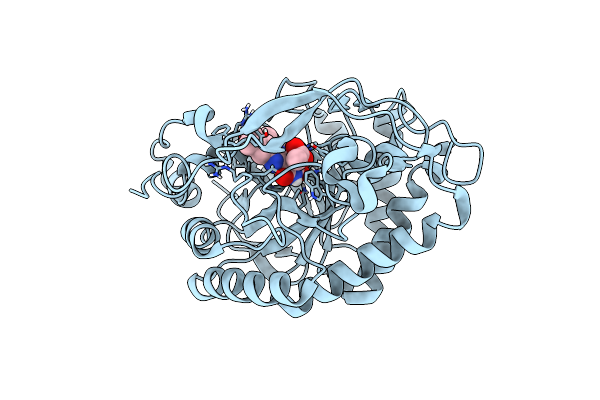 |
Crystal Structure Of Pentaerythritol Tetranitrate Reductase (Petnr) Mutant I107A
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-03-20 Classification: FLAVOPROTEIN Ligands: FMN, ACT |

