Search Count: 18
 |
Solution Structure Of The C65A/C167A Mutant Of Human Lipocalin-Type Prostaglandin D Synthase
|
 |
Organism: Canis lupus familiaris
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-11-10 Classification: ALLERGEN |
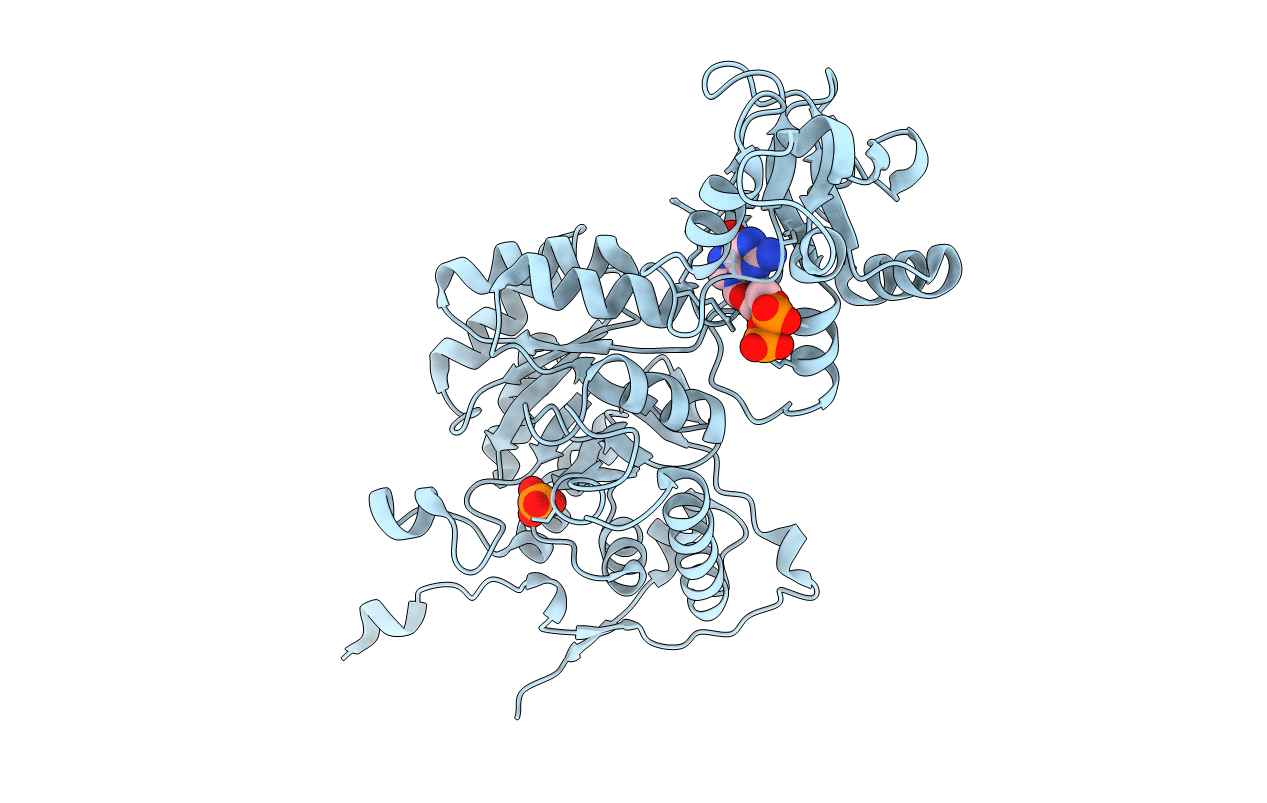 |
Crystal Structure Of Gmp Reductase From Trypanosoma Brucei In Complex With Guanosine 5'-Triphosphate
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: GTP, PO4 |
 |
Organism: Trypanosoma brucei brucei (strain iltat1.4)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-03-04 Classification: OXIDOREDUCTASE |
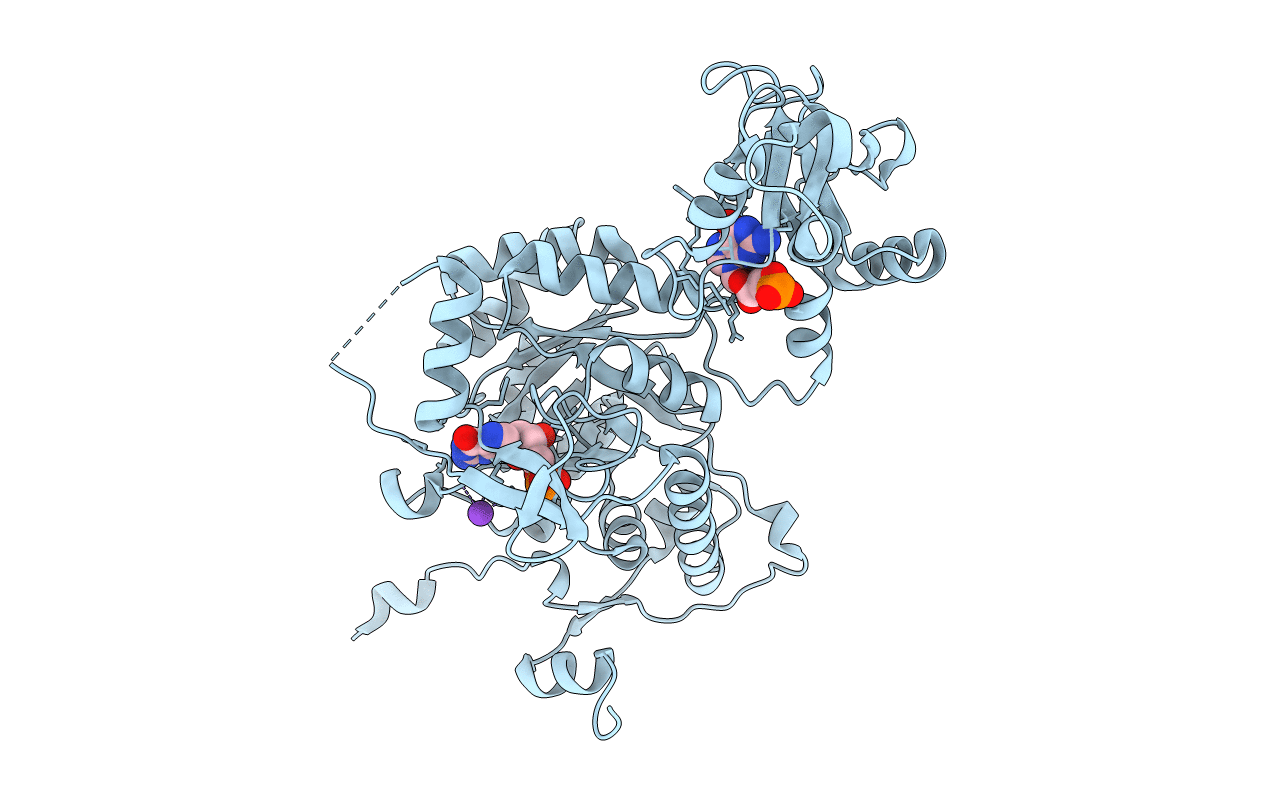 |
Crystal Structure Of Gmp Reductase C318A From Trypanosoma Brucei In Complex With Guanosine 5'-Monophosphate
Organism: Trypanosoma brucei brucei (strain iltat1.4)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-02-26 Classification: OXIDOREDUCTASE Ligands: 5GP, K |
 |
Organism: Canis lupus familiaris
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2018-04-04 Classification: ALLERGEN Ligands: PEG |
 |
Crystal Structure Of The Catalytic Domain Of Beta-Amylase From Paenibacillus Polymyxa
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-02-20 Classification: HYDROLASE Ligands: CA, CL, EDO, GOL, TRS, PEG |
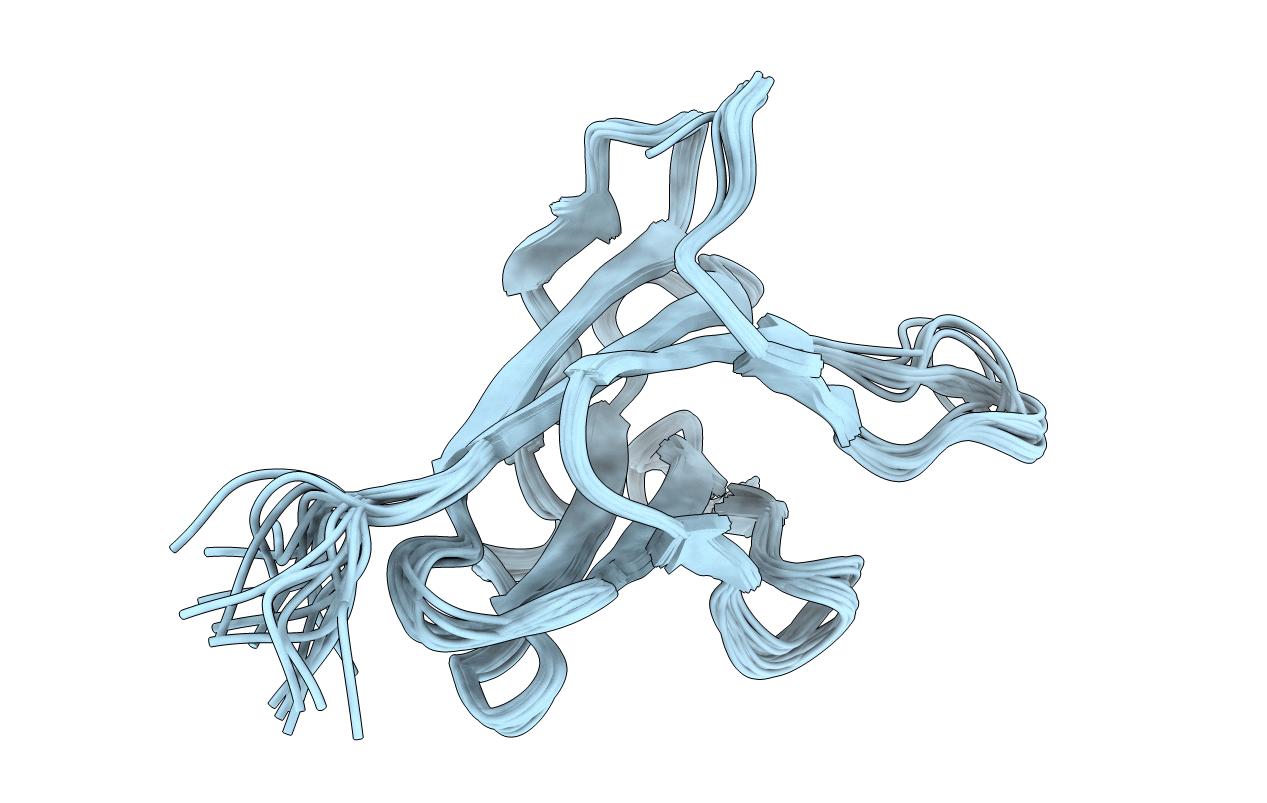 |
Solution Strucuture Of The Cbm25-1 Of Beta/Alpha-Amylase From Paenibacillus Polymyxa
Organism: Paenibacillus polymyxa
Method: SOLUTION NMR Release Date: 2012-04-04 Classification: HYDROLASE |
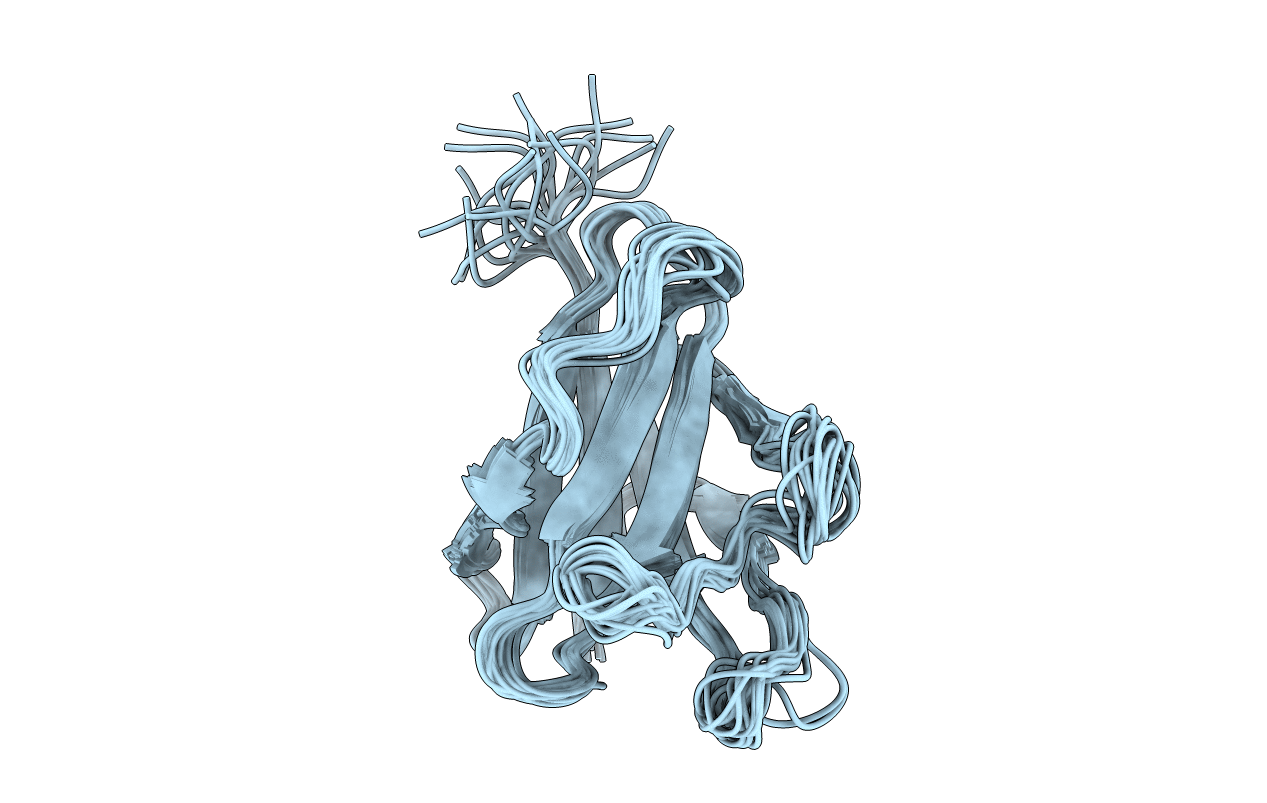 |
Solution Strucuture Of The Cbm25-2 Of Beta/Alpha-Amylase From Paenibacillus Polymyxa
Organism: Paenibacillus polymyxa
Method: SOLUTION NMR Release Date: 2012-04-04 Classification: HYDROLASE |
 |
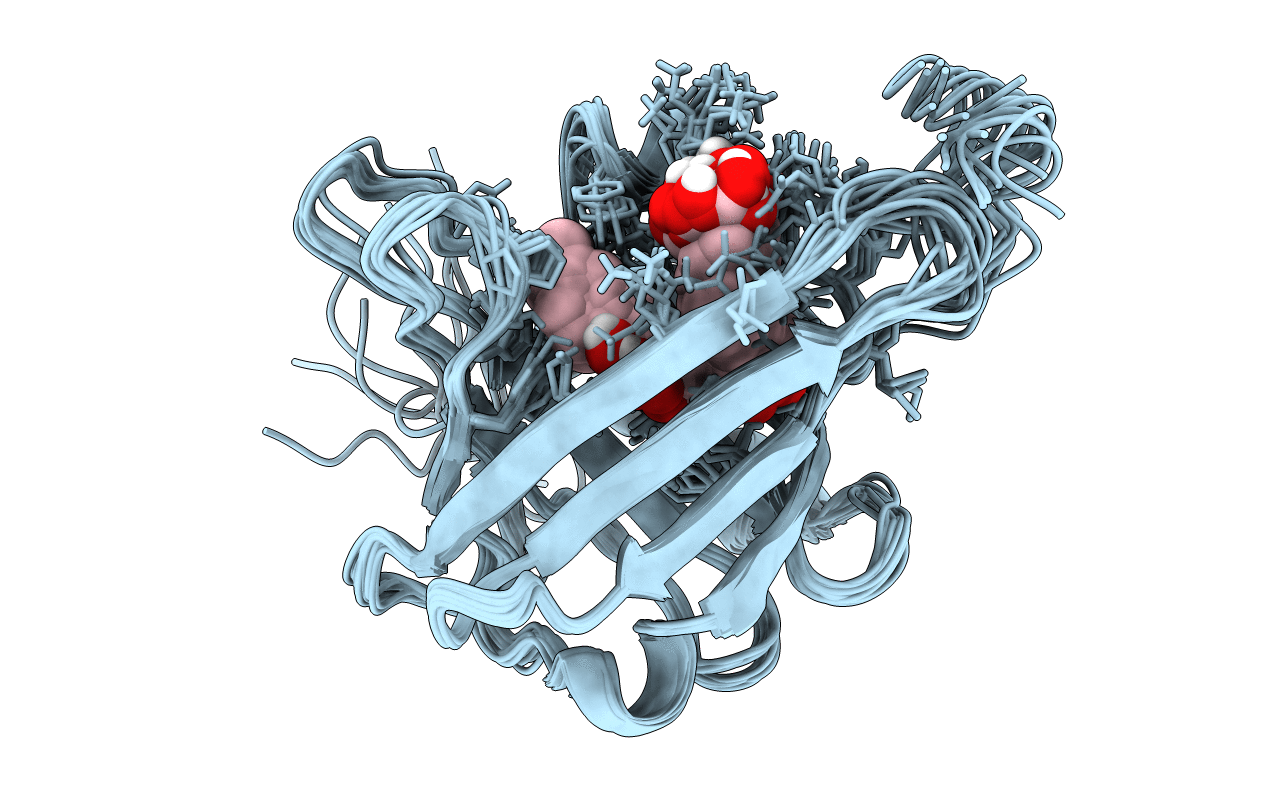
The 27-Residue N-Terminus Ccr5-Peptide In A Ternary Complex With Hiv-1 Gp120 And A Cd4-Mimic Peptide
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2011-07-27 Classification: SIGNALING PROTEIN |
 |
Solution Structure Of Mouse Lipocalin-Type Prostaglandin D Synthase / Substrate Analog (U-46619) Complex
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 2011-02-02 Classification: ISOMERASE Ligands: PUC |
 |
Crystal Structures Of Catalytic Site Mutants Of Active Domain 2 Of Thermostable Chitinase From Pyrococcus Furiosus Complexed With Chito-Oligosaccharides
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-06-09 Classification: HYDROLASE Ligands: MG, SO4 |
 |
Crystal Structures Of Catalytic Site Mutants Of Active Domain 2 Of Thermostable Chitinase From Pyrococcus Furiosus Complexed With Chito-Oligosaccharides
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2010-06-09 Classification: HYDROLASE Ligands: MG, SO4, GOL |
 |
Crystal Structures Of Catalytic Site Mutants Of Active Domain 2 Of Chitinase From Pyrococcus Furiosus
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2010-06-09 Classification: HYDROLASE Ligands: MG, GOL, SO4 |
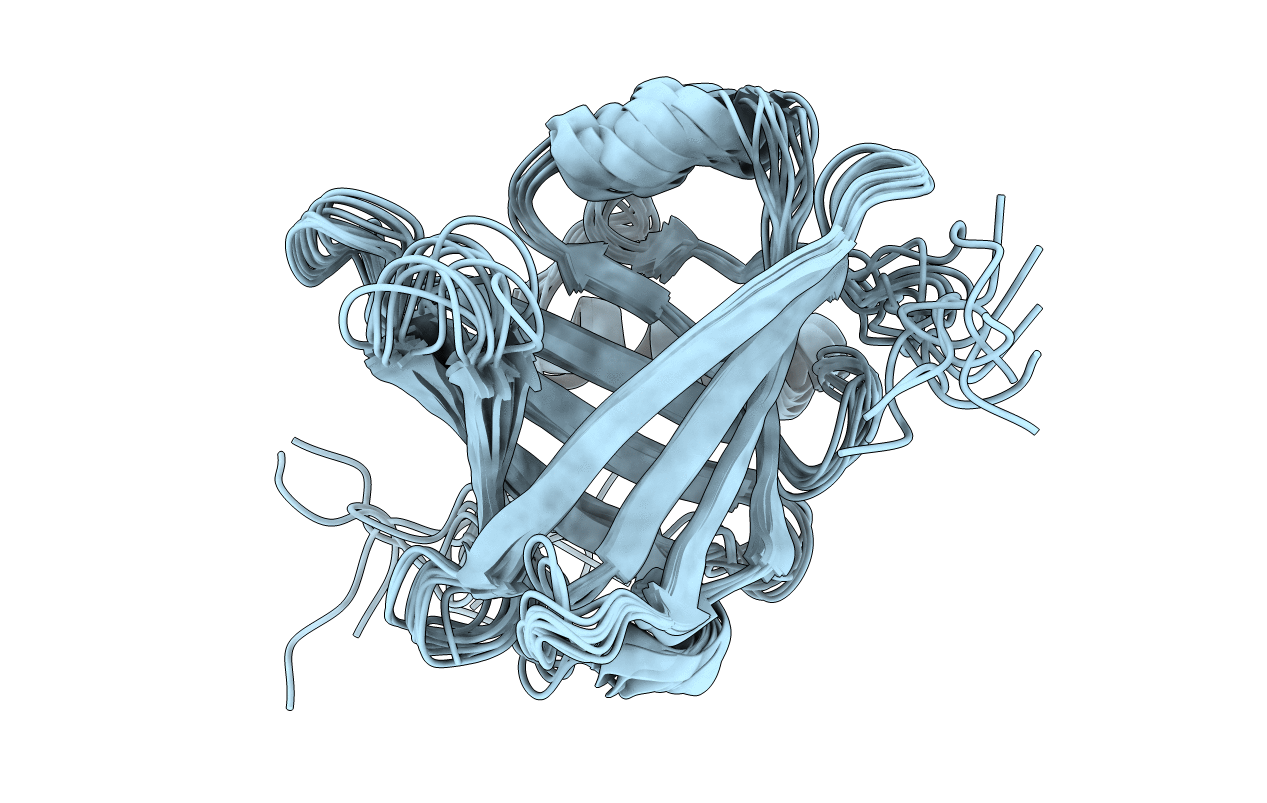 |
Solution Structure Of Mouse Lipocalin-Type Prostaglandin D Synthase Possessing The Intrinsic Disulfide Bond
|
 |
 |
 |

