Search Count: 14
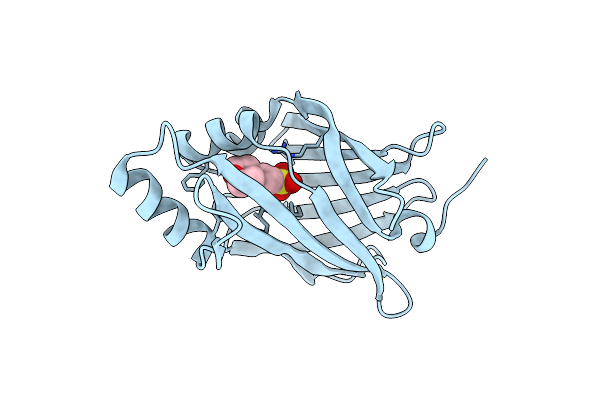 |
Crystal Structure Of Ql-Nanokaz (Reverse Mutant Of Nanokaz With L18Q And V27L)
Organism: Oplophorus gracilirostris
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-08-24 Classification: LUMINESCENT PROTEIN Ligands: MES |
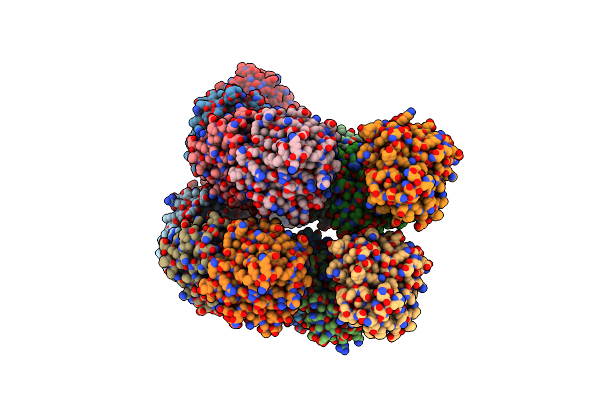 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2021-06-23 Classification: OXIDOREDUCTASE Ligands: J2X |
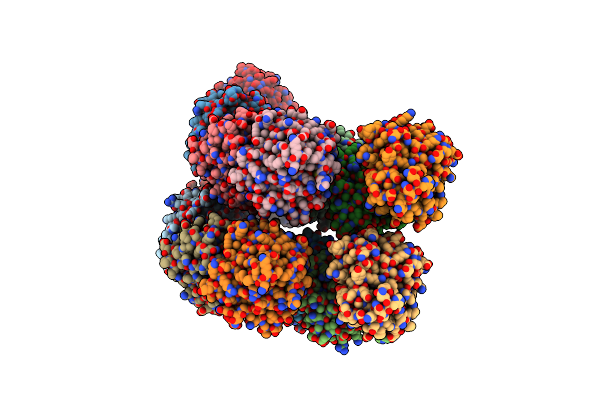 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2021-06-23 Classification: OXIDOREDUCTASE Ligands: J2U |
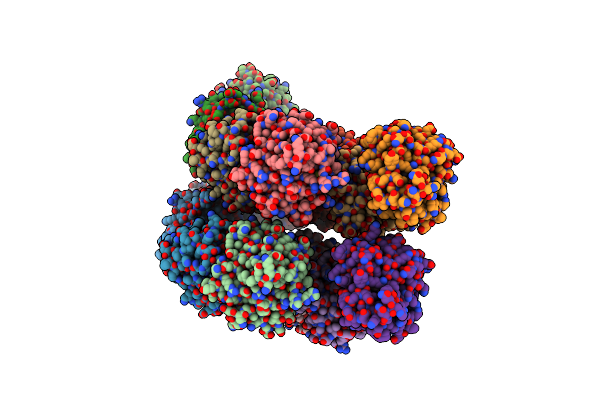 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-06-06 Classification: LUMINESCENT PROTEIN Ligands: 9A3 |
 |
Crystal Structure Of The Mutated 19 Kda Protein Of Oplophorus Luciferase (Nanokaz)
Organism: Oplophorus gracilirostris
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2016-01-13 Classification: LUMINESCENT PROTEIN |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2005-02-08 Classification: LUMINESCENT PROTEIN Ligands: CZP |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2005-02-08 Classification: LUMINESCENT PROTEIN Ligands: CZI |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2005-02-08 Classification: LUMINESCENT PROTEIN Ligands: CZB |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2005-02-08 Classification: LUMINESCENT PROTEIN Ligands: CZN |
 |
Organism: Aequorea aequorea
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2000-05-31 Classification: OXIDOREDUCTASE Ligands: CZH |
 |
Nmr-Derived Three-Dimensional Solution Structure Of Protein S Complexed With Calcium
Organism: Myxococcus xanthus
Method: SOLUTION NMR Release Date: 1994-08-31 Classification: BINDING PROTEIN Ligands: CA |
 |
Nmr-Derived Three-Dimensional Solution Structure Of Protein S Complexed With Calcium
Organism: Myxococcus xanthus
Method: SOLUTION NMR Release Date: 1994-08-31 Classification: BINDING PROTEIN Ligands: CA |
 |
Crystal Structure Of Myxococcus Xanthus Nucleoside Diphosphate Kinase And Its Interaction With A Nucleotide Substrate At 2.0 Angstroms Resolution
Organism: Myxococcus xanthus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1994-05-31 Classification: PHOSPHOTRANSFERASE(PO4 AS ACCEPTOR) Ligands: MG, ADP |
 |
Crystal Structure Of Myxococcus Xanthus Nucleoside Diphosphate Kinase And Its Interaction With A Nucleotide Substrate At 2.0 Angstroms Resolution
Organism: Myxococcus xanthus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1994-01-31 Classification: PHOSPHOTRANSFERASE(PO4 AS ACCEPTOR) |

