Search Count: 46
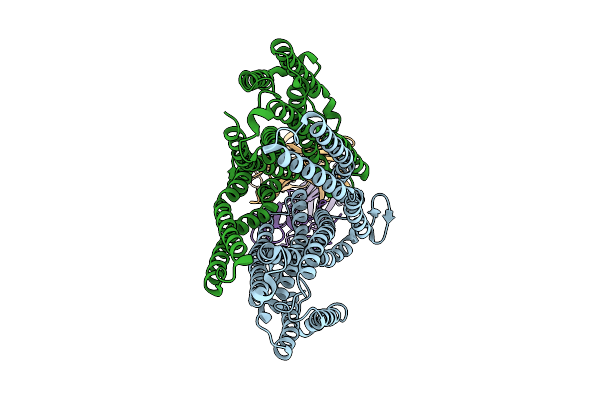 |
Cryoem Structure Of The Proton-Dependent Antibacterial Peptide Transporter Sbma In Complex With Fabs11-1 In Lipid Nanodiscs At Ph 5.5, Inward-Open State
Organism: Escherichia coli, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: TRANSPORT PROTEIN |
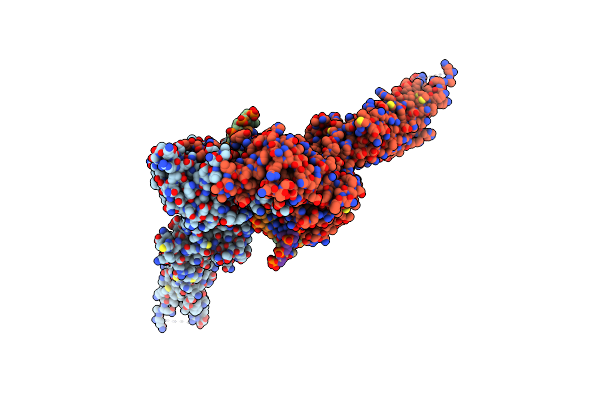 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.84 Å Release Date: 2025-03-26 Classification: IMMUNE SYSTEM |
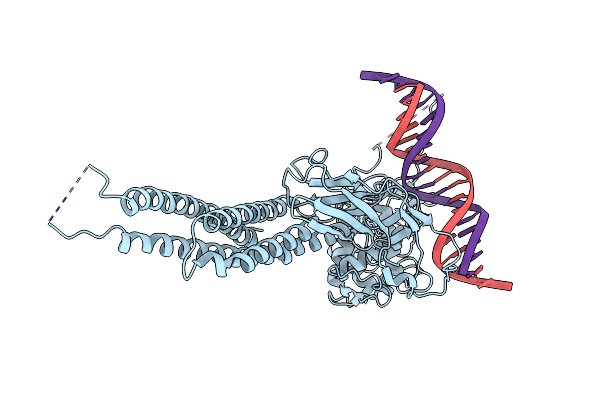 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.07 Å Release Date: 2025-03-26 Classification: IMMUNE SYSTEM |
 |
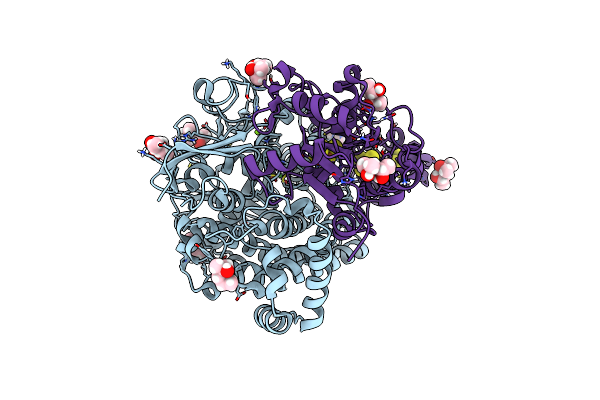
Organism: Microcystis aeruginosa nies-88
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-06-19 Classification: TRANSFERASE Ligands: PIS, MG, BTB |
 |
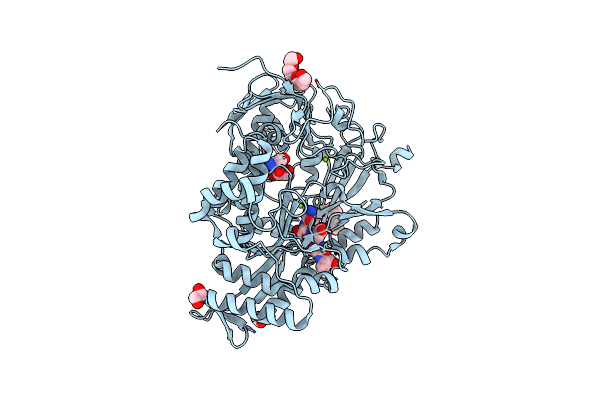
Neutron Structure Of [Nife]-Hydrogenase From D. Vulgaris Miyazaki F In Its Oxidized State
Organism: Desulfovibrio vulgaris str. 'miyazaki f'
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.04 Å, 2.20 Å Release Date: 2023-09-13 Classification: OXIDOREDUCTASE Ligands: NFU, MPD, OH, MG, CL, SF4, F3S |
 |
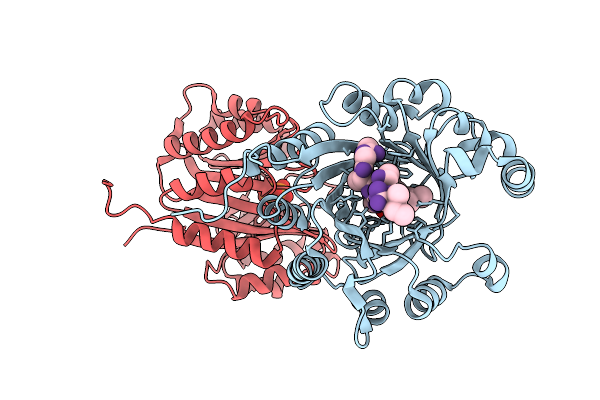
Crystal Structure Of The Substrate-Binding Protein Yeja From S. Meliloti In Complex With Peptide Fragment
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2023-02-22 Classification: TRANSPORT PROTEIN Ligands: PG4, EDO, MG |
 |
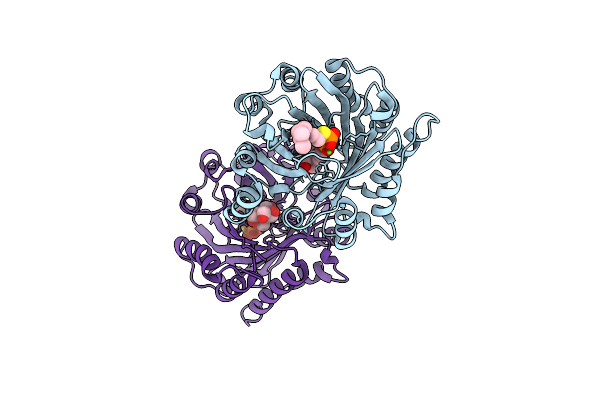
Crystal Structure Of Limf Prenyltransferase Bound With A Peptide Substrate And Gspp
Organism: Limnothrix sp. caciam 69d, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2022-08-03 Classification: TRANSFERASE Ligands: MG, GST, PPV |
 |
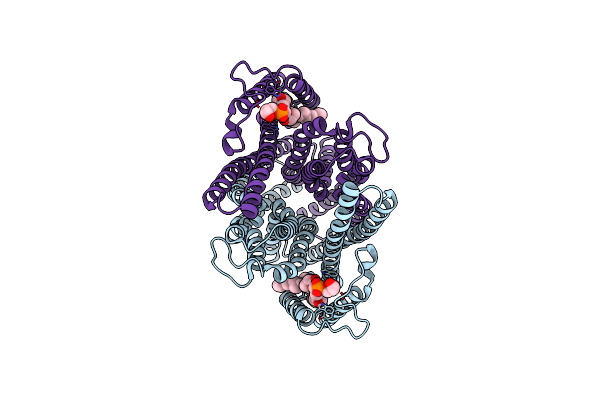
Organism: Limnothrix sp. caciam 69d
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2022-08-03 Classification: TRANSFERASE Ligands: MG, GST, BTB |
 |
Cryo-Em Structure Of The Proton-Dependent Antibacterial Peptide Transporter Sbma-Fabs11-1 In Nanodiscs
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-09-15 Classification: TRANSPORT PROTEIN Ligands: PGW |
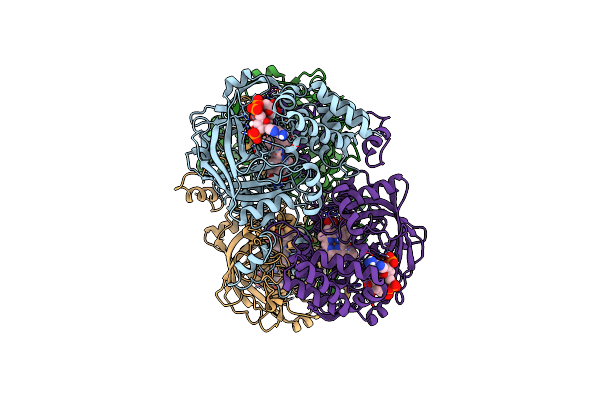 |
Electron Crystallographic Structure Of Catalase Using A Direct Electron Detector At 300 Kv
Organism: Bos taurus
Method: ELECTRON CRYSTALLOGRAPHY Resolution:3.20 Å Release Date: 2020-12-09 Classification: OXIDOREDUCTASE Ligands: HEM, NDP |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2017-12-20 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-12-06 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2017-12-06 Classification: HYDROLASE |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2017-11-29 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Engyodontium album
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-11-29 Classification: HYDROLASE Ligands: NO3, PR |
 |
Organism: Engyodontium album
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2017-06-07 Classification: HYDROLASE Ligands: CA, NO3 |
 |
Organism: Engyodontium album
Method: X-RAY DIFFRACTION Resolution:0.98 Å Release Date: 2017-06-07 Classification: HYDROLASE Ligands: NO3, GOL, CA |
 |
 |
 |

